Isolated DNA from the remaining ctenidia tissue samples from Ronit’s experiment (Google Sheet).
Tissue was excised from frozen tissue block via razor blade (weight not recorded) and pulverized under liquid nitrogen. Samples were incubated O/N @ 37oC (heating block) in 350uL of MB1 Buffer + 25uL Proteinase K, per the E.Z.N.A. Mollusc DNA Kit (Omega) instructions.
After the O/N incubation, I processed the samples according to the E.Z.N.A. Mollusc DNA Kit (Omega) with the following notes:
Samples were eluted in 150uL of Elution Buffer.
Samples were stored in “Ronit’s gDNA Box #1 (positions A1 - B8 and C1 - E4)” in the FTR213 -20oC freezer.
See either of the spreadsheets for the full list of samples isolated today.
NOTE: One sample (D18) did not isolate properly. After the chloroform:IAA treatment, the sample did not produce an aqueous phase (even after the specified troubleshooting step in the manual). I will have to perform another isolation on this sample next week.
Here’s an image of how the sample appeared:
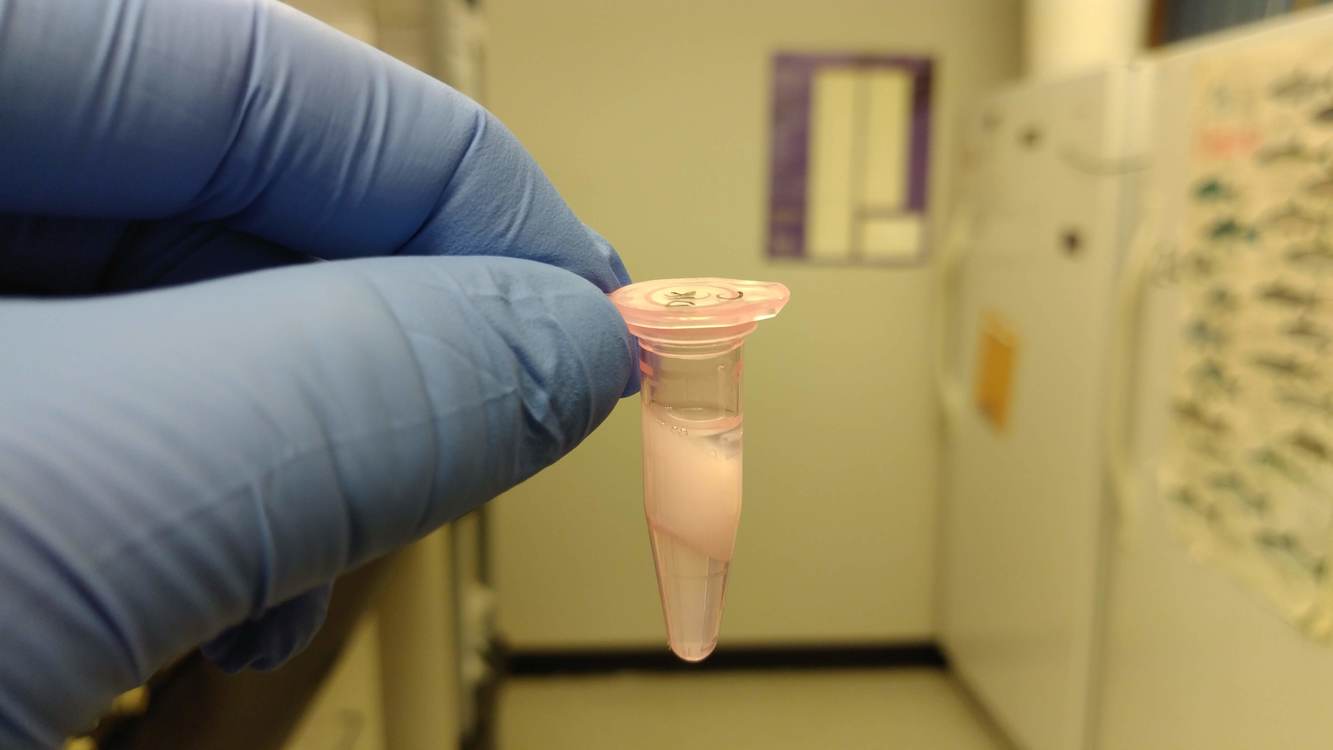
That white “fluffy” interphase should be highly compressed in a very tight, flat, smooth layer separating the organic (bottom clear liquid) and the aqueous (top clear liquid) phases.