I isolated a bunch of tanner crab RNA on 20190430 and Steven asked me to try to figure out some options for RNAseq libray pools.
Using Grace’s table join of my Qubit data (hemo_qub-for-libs.csv), I created a pivot table to quickly get an idea of how things looked (note: I added a column with total RNA yield for each sample - 10uL * concentration):
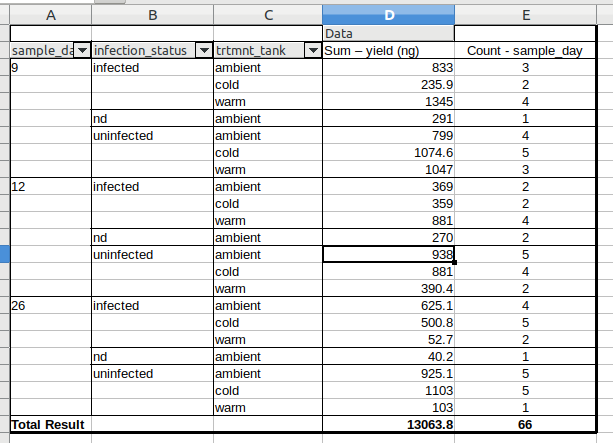
Assuming a minimum of 1000ng is required by the UW’s Northwest Genomics Center (that was the requirement last time we had RNAseq performed by them; have emailed to confirm it’s still required), here are the libraries I think we can/should make.
Possible libraries, by “Day”:
Day 9
- Infected vs Uninfected
Day 12
- Infected vs Uninfected
- Ambient vs Cold vs Warm
Day 26
- Infected vs Uninfected
- Ambient vs Cold
The “Day 9” samples are named poorly, as these are actually the samples prior to temperature exposures. Thus, the “trtmnt_tank” column (i.e. temperature treatment) in the table above doesn’t really apply. With that in mind, Day 9 samples can only be compared as Infected/Uninfected.
So, by default, those should be two libraries.
Then, we have the Day 12 and Day 26 samples. We can either do Infected/Uninfected OR Ambient vs. Cold, but can’t do both, as there isn’t sufficient RNA…
Admittedly, I’m not sure which aspect of this project was given more weight in the grant proposal:
- response to disease over time
- response to temperature over time
The answer to that question will guide library pooling.
With that said, if we do the following, we’ll have a bit of both pathogen response and temperature response:
Day 9 Infected/Uninfected
Day 12 Ambient/Cold
Day 26 Ambient/Cold