We recently acquired a NanoPore MinION sequencer, FLO-MIN106 flow cell and the Rapid Sequencing Kit (SQK-RAD004). The NanoPore website provides a pretty thorough an user-friendly walk-through of how to begin using the system for the first time. With that said, I believe the user needs to have a registered account with NanoPore and needs to have purchased some products to have full access to the protocols they provide.
For first time users, they provide a “Lambda Control experiment” which:
teaches you how to use the sequencer, flow cells, and sequencing Kit
sequences a known, small genome to allow fast sequencing and analysis
free access to their
EPI2MEanalysis platform (which will perform basecalling, quality analysis, alignment, and generate a visually pleasing sequencing summary/report)
Honestly, it’s a really helpful and easy way to get introduced to using the entire system. It’s also not a bad way to sell the EPI2ME service, as it’s very hands-off and easy to run.
Anyway, I set up the Lambda Control experiment sequencing run and ran it for the recommended duration (6hrs) with basecalling enabled (this will help speed up the subsequent EPI2ME service after sequencing is complete).
RESULTS
Output folder:
Fast5 format reads:
FastQ format reads:
EPI2ME Report #1 (Flow cell performance; PDF):
EPI2ME Report #2 (Alignment stats; PDF):
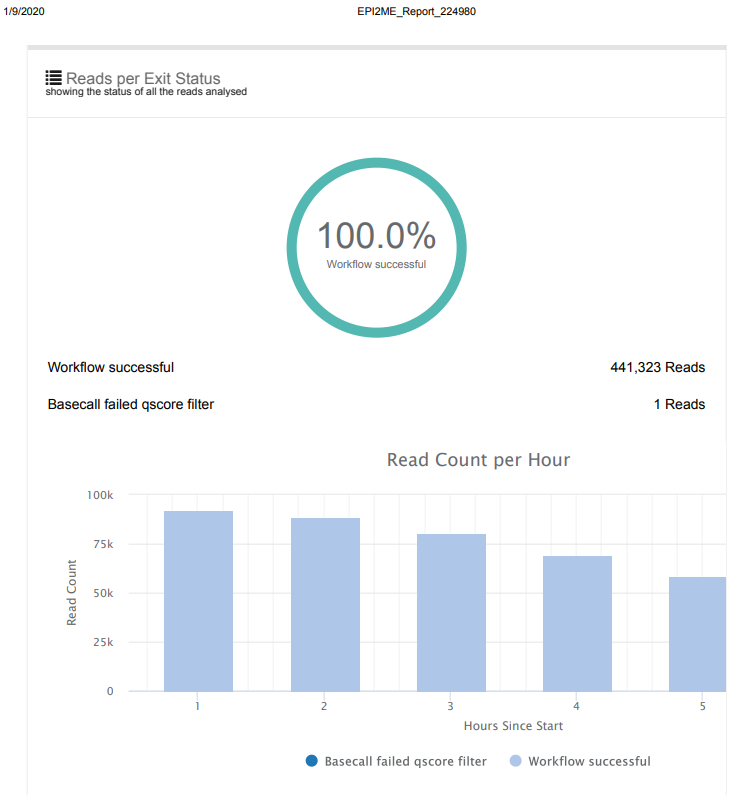
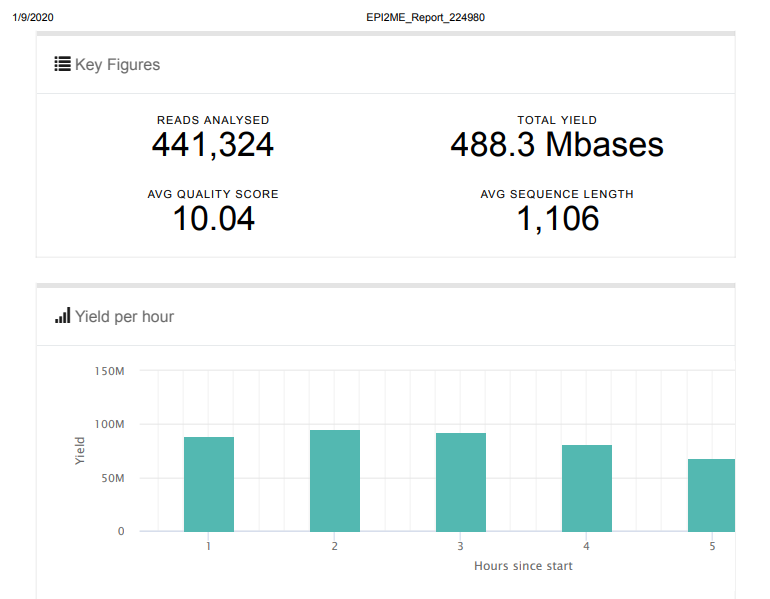
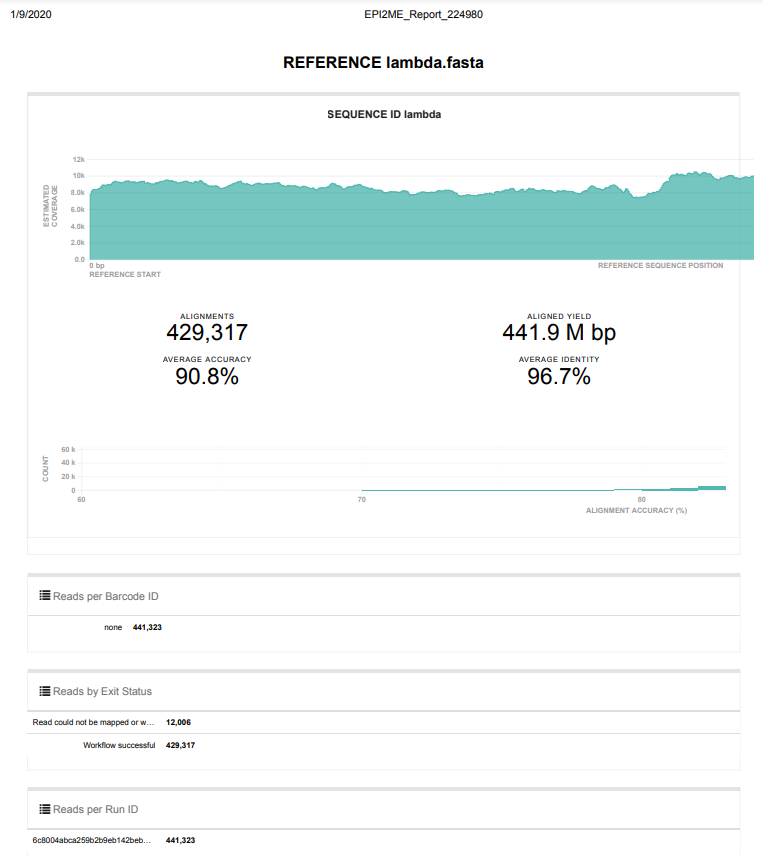
Overall, the run went as expected and yielded >8,000x coverage of the Lambda genome, with only ~2.8% of reads failing to map to the genome. Will proceed to running an actual sample!
Screencaps below are taken directly from the two EPI2ME reports linked above.