After completing annotation of the C.bairdi MEGAN6 taxonomic-specific Trinity assembly using Trinotate on 20200126, I performed differential gene expression analysis and gene ontology (GO) term enrichment analysis using Trinity’s scripts to run EdgeR and GOseq, respectively, across all of the various treatment comparisons. The comparison are listed below and link to each individual SBATCH script (GitHub) used to run these on Mox.
It should be noted that most of these comparisons do not have any replicate samples (e.g. D12 infected vs D12 uninfected). I made a weak attempt to coerce some results from these by setting a dispersion value in the edgeR command. However, I’m not expecting much, nor am I certain I would really trust the results from those particular comparisons.
RESULTS
Output folder:
Comparisons:
D12_infected-vs-D12_uninfected
Took a little less than 20mins to run:

Only a single DEG, which is upregulated in the infected set:
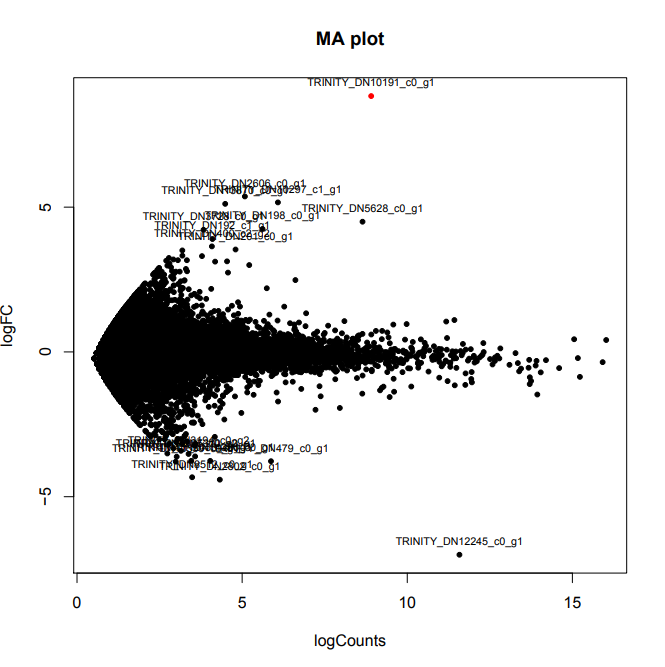
TRINITY_DN10191_c0_g1 - SPID: Q36421 (Cyctochrome c oxidase I)
D12_infected-vs-D26_infected
Took ~18mins to run:

No differentially expressed genes between these two groups.
NOTE: Since no DEGs, that’s why this run shows as FAILED in the above runtime screencap. This log file captures the error message that kills the job and generates the FAILED indicator:
Error, no differentially expressed transcripts identified at cuttoffs: P:0.05, C:1 at /gscratch/srlab/programs/trinityrnaseq-v2.9.0/Analysis/DifferentialExpression/analyze_diff_expr.pl line 203.
D12_uninfected-vs-D26_uninfected
Took ~18mins to run:

No differentially expressed genes between these two groups.
NOTE: Since no DEGs, that’s why this run shows as FAILED in the above runtime screencap. This log file captures the error message that kills the job and generates the FAILED indicator:
Error, no differentially expressed transcripts identified at cuttoffs: P:0.05, C:1 at /gscratch/srlab/programs/trinityrnaseq-v2.9.0/Analysis/DifferentialExpression/analyze_diff_expr.pl line 203.
D12-vs-D26
Took ~40mins to run:

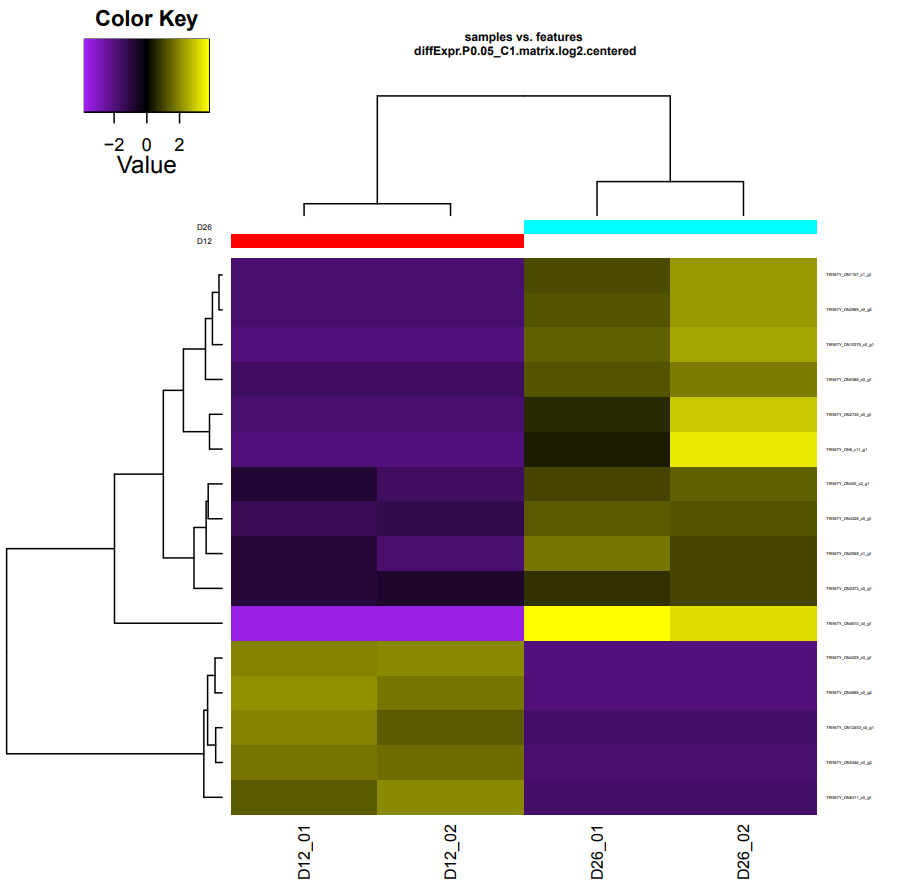
D12 upregulated genes:
Five genes:
TRINITY_DN4239_c0_g1 - No annotation
TRINITY_DN4669_c0_g2 - No annotation
TRINITY_DN5346_c0_g2 - No annotation
TRINITY_DN12453_c0_g1 - SP ID: Q6ING4(DEP domain-containing protein 1A)
TRINITY_DN8311_c0_g1 - No annotation
D12 GO enrichment identified zero enriched and five depleted:
-
- Only one of these five is in the “biological process” category and it is uncharacterized (i.e. is identified as “biological process”).
D26 upregulated genes:
11 genes:
TRINITY_DN4610_c0_g1 - SP ID: Q9MFN9(Cytochrome b)
TRINITY_DN10370_c0_g1 - SP ID: P20241(Neuroglian)
TRINITY_DN2559_c1_g1 - No annotation.
TRINITY_DN5386_c0_g1 - No annotation.
TRINITY_DN400_c2_g1 - SP ID: Q8N587(Zinc finger protein 561)
TRINITY_DN2969_c0_g2 - No annotation.
TRINITY_DN4328_c0_g1 - No annotation.
TRINITY_DN8_c11_g1 - No annotation.
TRINITY_DN1107_c1_g1 - No annotation.
TRINITY_DN2373_c0_g1 - SP ID: Q4AEI0(Glutathione peroxidase 2)
TRINITY_DN2730_c0_g1 - No annotation.
D26 GO enrichment identified four up-regulated enriched GO terms (all in the “molecular function” category) and five up-regulated depleted GO terms (all in the “biological process” category).
D26_infected-vs-D26_uninfected
No differentially expressed genes between these two groups.
NOTE: Since no DEGs, that’s why this run shows as FAILED in the above runtime screencap. This log file captures the error message that kills the job and generates the FAILED indicator:
20200207_cbai_DEG/D26_infected-vs-D26_uninfected/edgeR.20733.dir/diff_expr_stderr.txt
Error, no differentially expressed transcripts identified at cuttoffs: P:0.05, C:1 at /gscratch/srlab/programs/trinityrnaseq-v2.9.0/Analysis/DifferentialExpression/analyze_diff_expr.pl line 203.
infected-vs-uninfected
Took ~40mins to run:

Output folder:
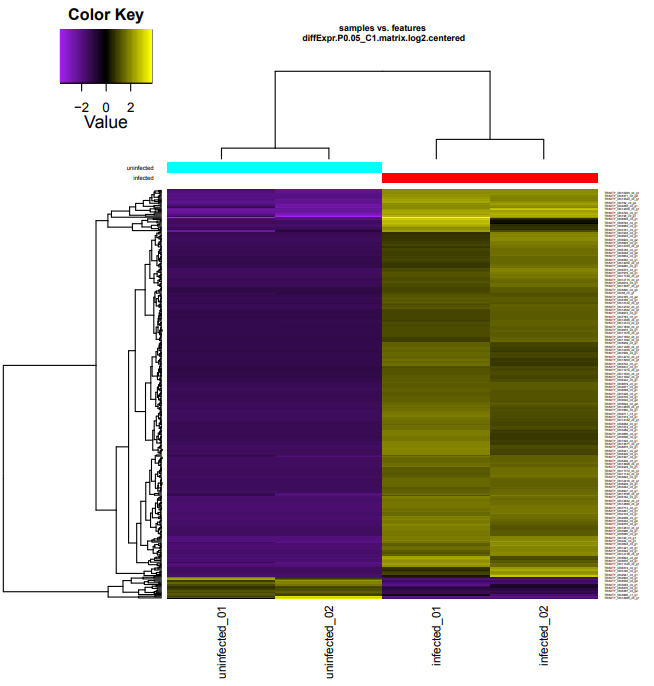
Infected upregulated DEGs:
Infected GO enrichment identified 374 enriched GO terms:
Uninfected upregulated genes:
Uninfected GO enrichment identified zero enriched GO terms.