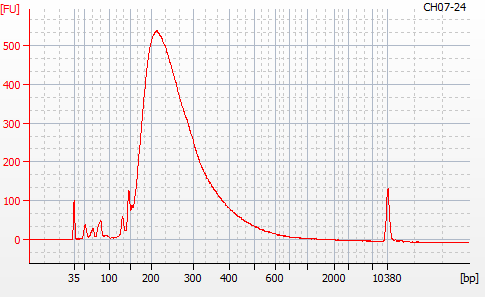
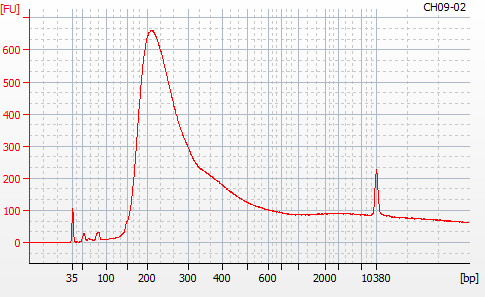
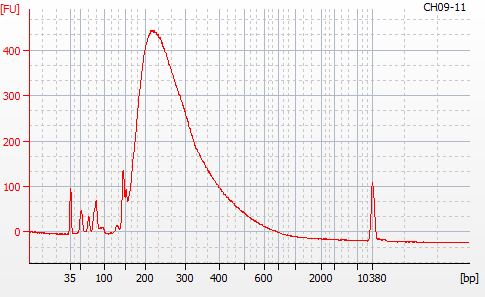
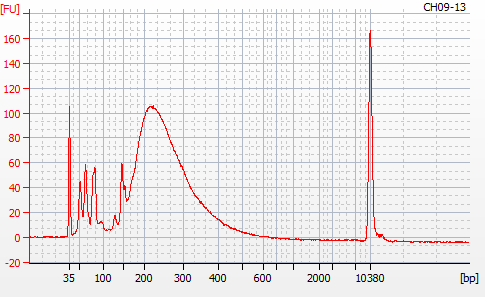
MBD BSseq library construction was completed yesterday (20201124). Next, I needed to evaluate the libraries using the Roberts Lab Bioanalyzer 2100 (Agilent) to assess library sizes, yields, and qualities (i.e. primer dimers).
ZymoResearch recommends using the TapeStation (Agilent), but if a lab doesn’t have access to that, they indicated that either of the regular DNA assays will work (verbal communication). I ran the libraries on the High Sensitivity DNA Assay (Agilent), since that’s what we had in the lab. It should work relatively OK.
All data (including Bioanalyzer electropherograms) will be added to the Google Sheet for this project:
Additional details are available in this GitHub repo:
RESULTS
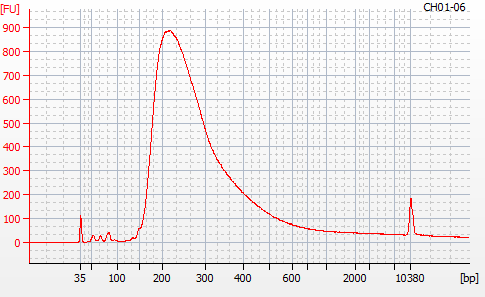
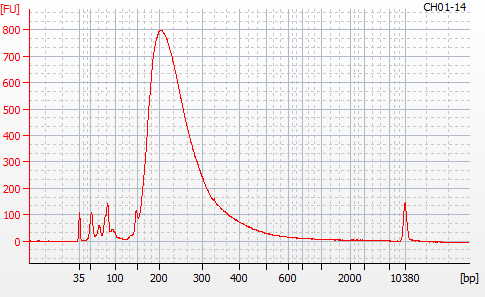
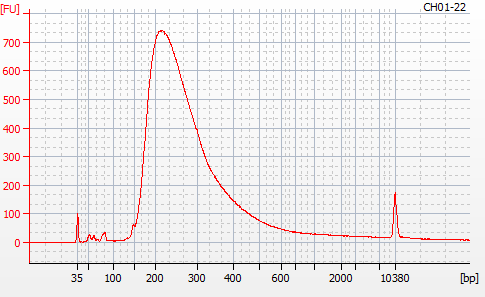
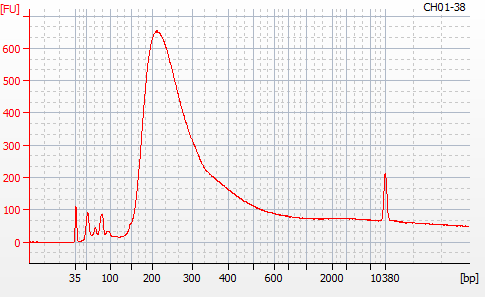
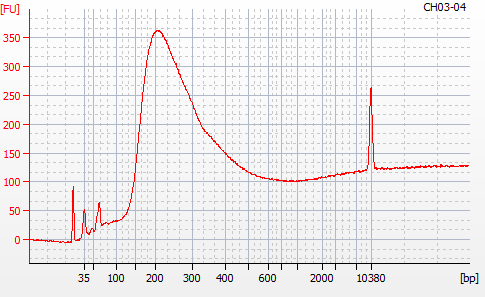
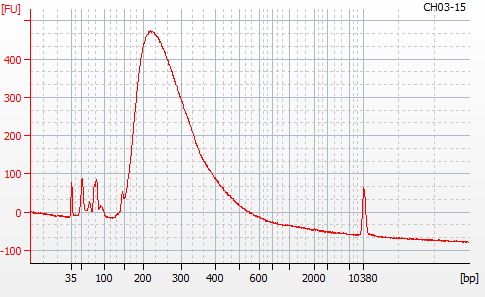
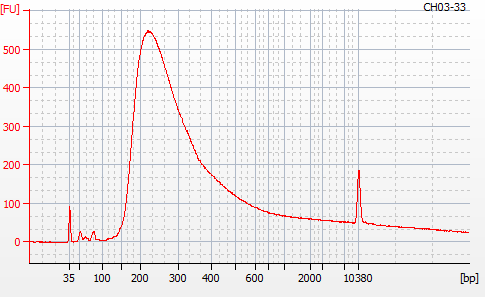
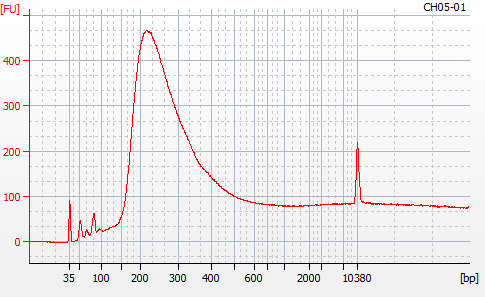
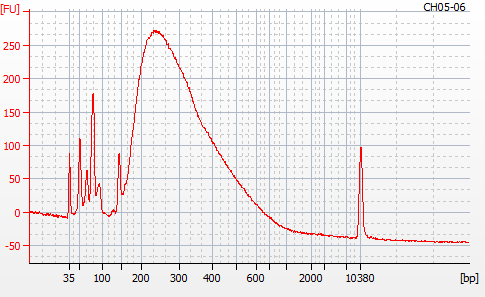
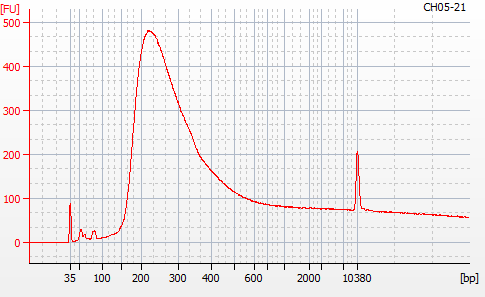
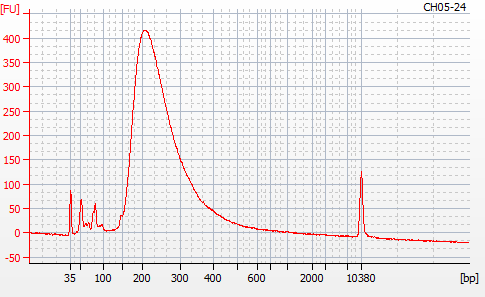
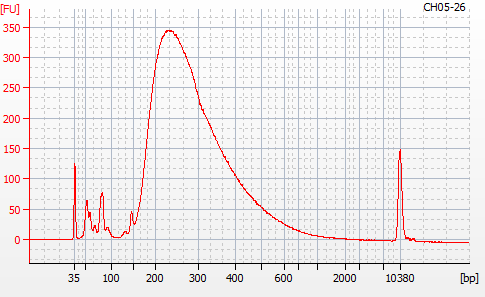
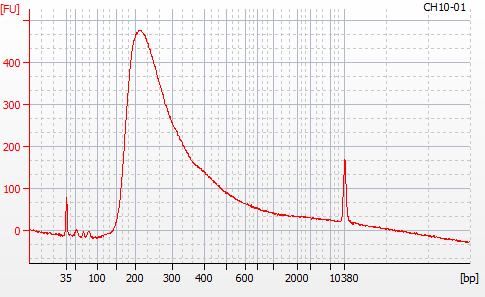
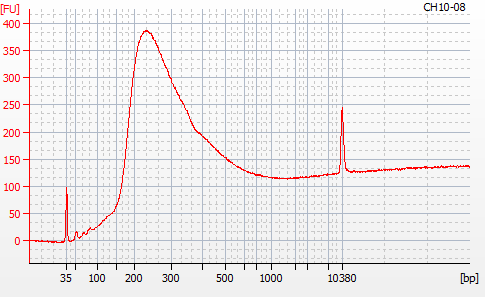
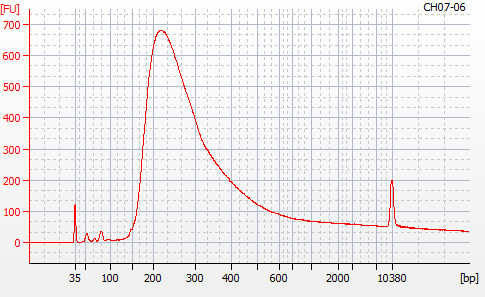
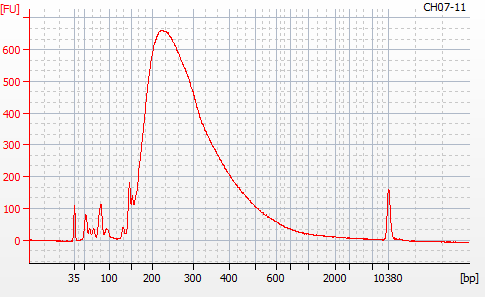
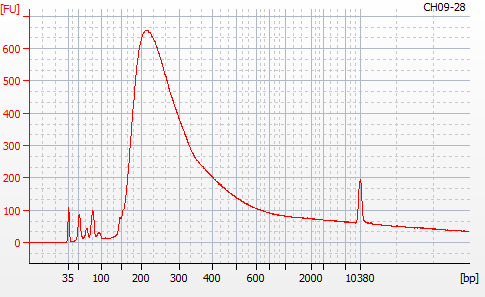
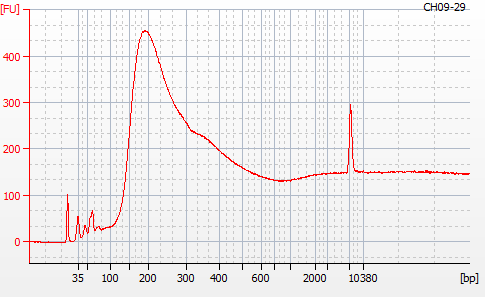
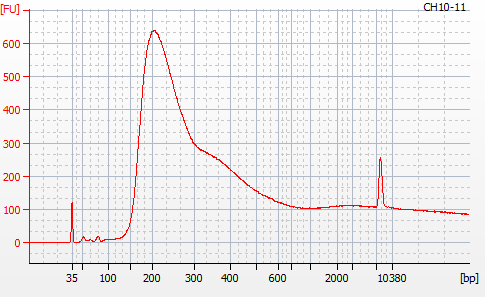
Most samples ([including CH09-13 which had no detectable DNA via Qubit(https://robertslab.github.io/sams-notebook/posts/2020/2020-11-03-MBD-Selection—M.magister-Sheared-Gill-gDNA-16-of-24-Samples-Set-3-of-3/)]) exhibited the expected profiles. Admittedly, a number of samples were probably slightly too concentrated for the High Sensitivity DNA Assay (Agilent), leading to skewed baselines. However, the software does a good job of detecting, and accounting for, this.
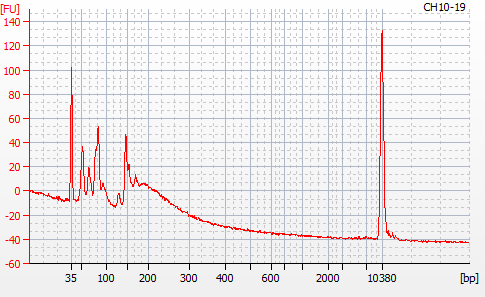
One sample failed: CH10-19
Although I did not expect it to fail, this sample did have a wonky elution at the end of SECTION 4 of the library prep..
For generating Bioanalyzer reports, a region was set for each sample from 44.52s to 109.57s (corresponding to 50bp and 8000bp, respecctively). A couple of samples had to be slightly adjusted due to the software having difficulty automatically identifying the lower marker. In the reports, each sample has a “Region Table” section and a corresponding “Region 1” which lists the following data (most are self-explanatory; the Color is simply the color selected to define “Region 1” on the electropherograms):
From [s]To [s]Corr. Area% of TotalAverage Size [bp]Size distribution in CV [%]Conc. [pg/ul]Molarity [pmol/l]Color
Once a sequencing facility is decided upon, will make decisions about sample pooling and desired sequencing output.
Output folder:
20201125_mmag_bioanalyzer_mbd-bsseq-libraries/
Bioanalyzer data files (XAD; requires 2100 Expert software):
Bioanalyzer report files (includes fragment lengths, concentration, and molarity; CSV):
Electropherograms and gels (full chips)
DE72902486_2020-11-25_05-16-07
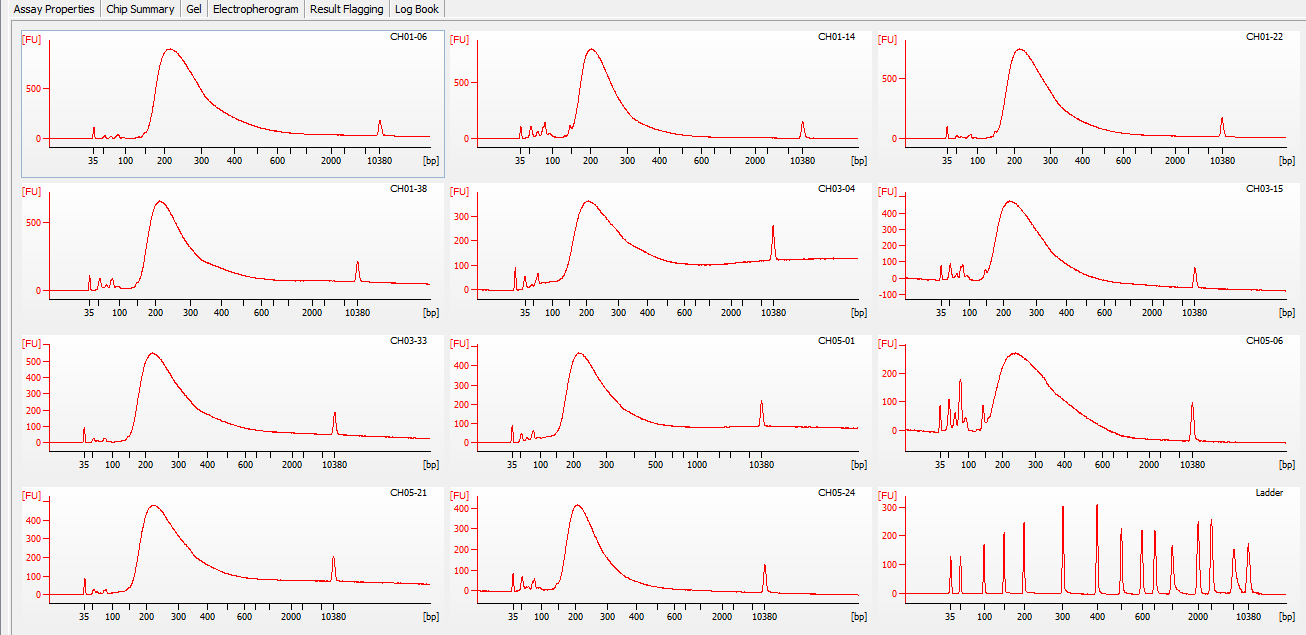
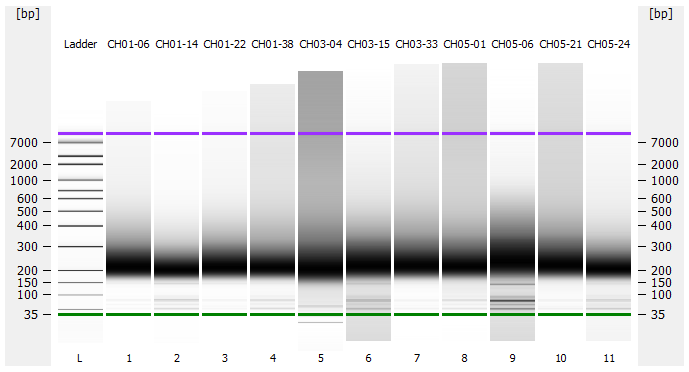
DE72902486_2020-11-25_06-12-38
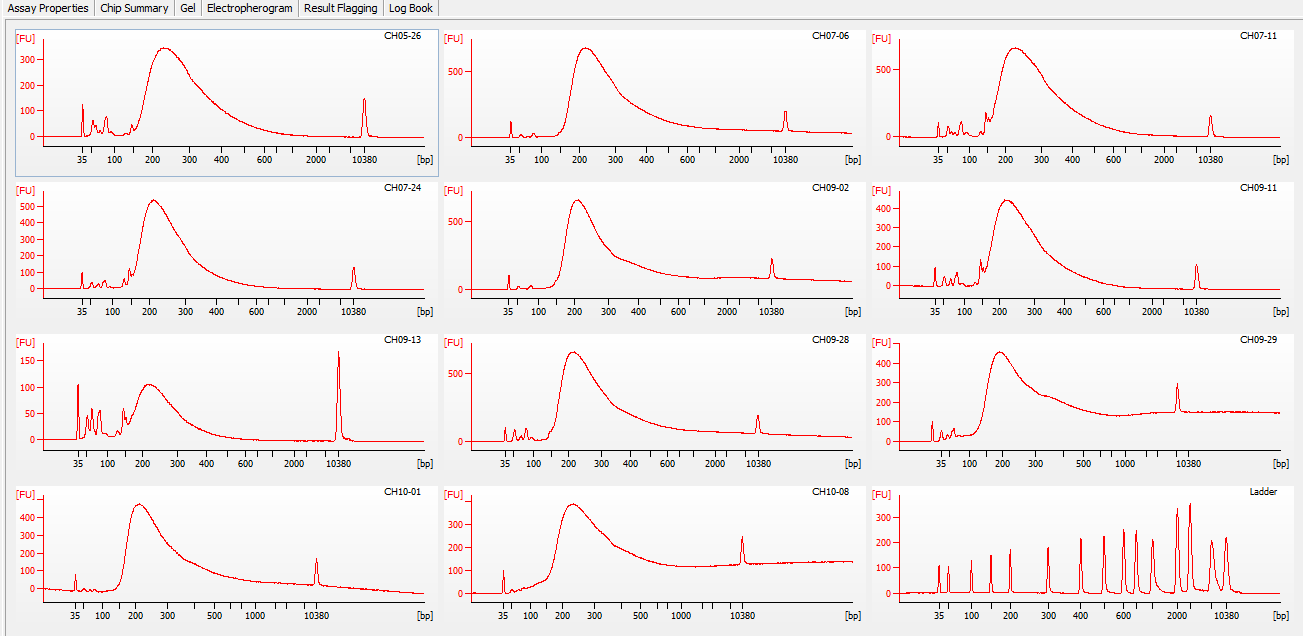
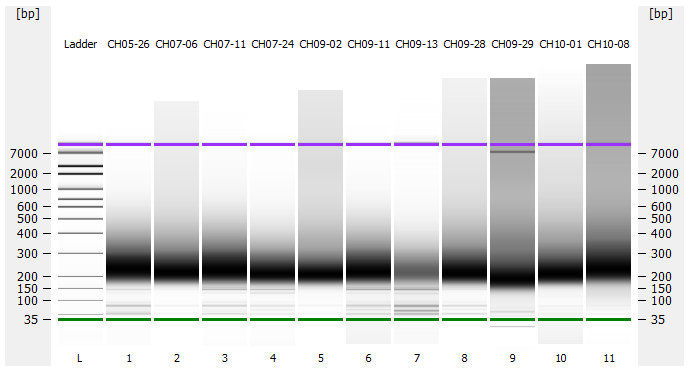
DE72902486_2020-11-25_07-00-25
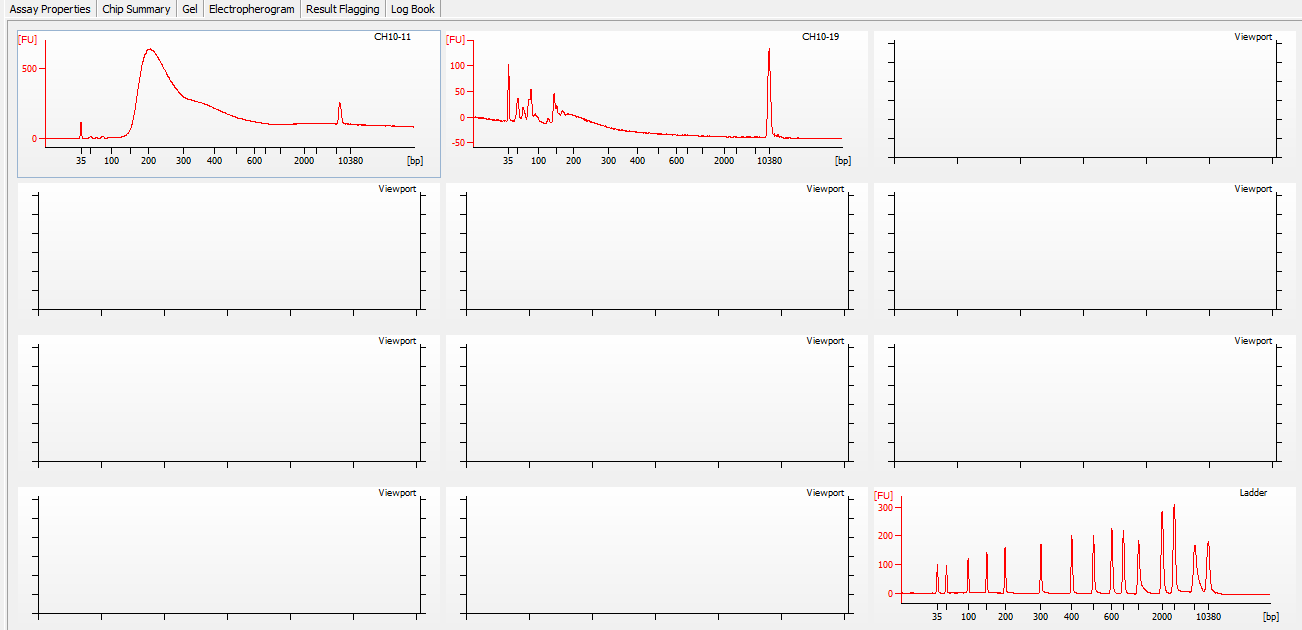
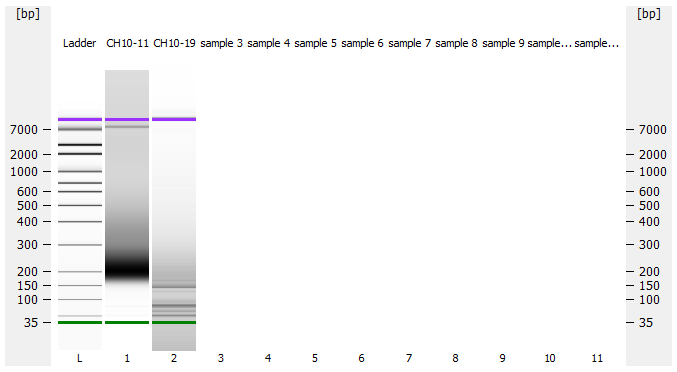
Electropherograms (individual samples)