Per this GitHub Issue, Steven wanted me to download all of the SRA data (RNA-seq and WGBS-seq) from NCBI BioProject PRJNA744403 and run QC on the data.
Prior to QC, I had to get the data:
Acquired accessions list file:
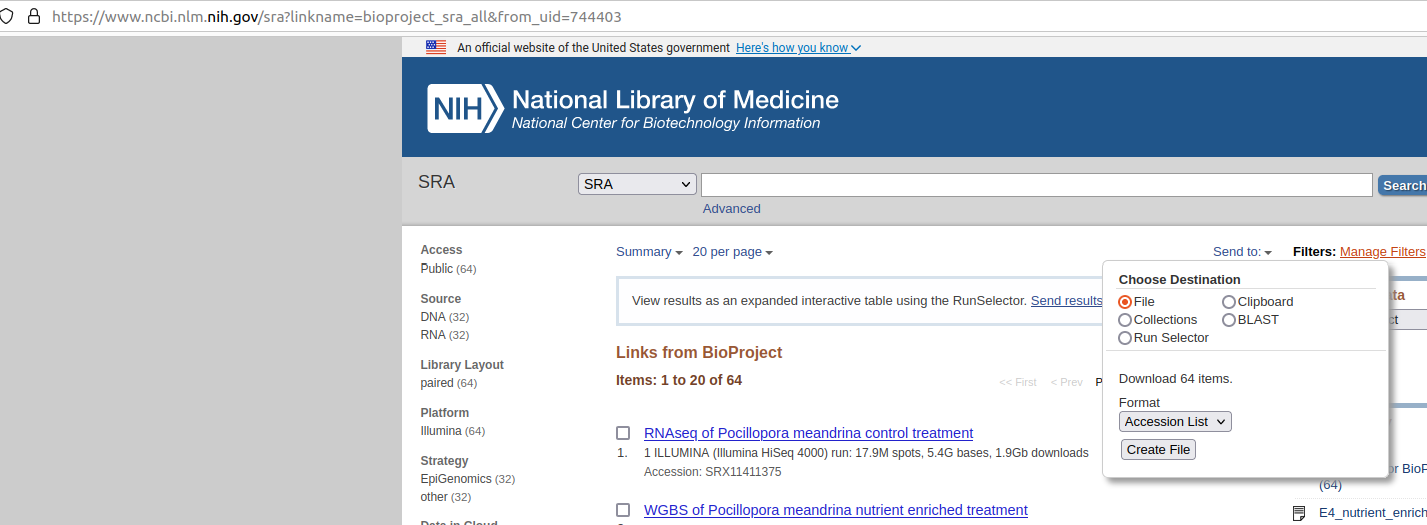
Screenshot of NCBI BioProject PRJNA744403 page showing dropdown menu to generate SRA accessions list file Resulting file looks like this:
Run accession SRR15101687 SRR15101688 SRR15101689 SRR15101690 SRR15101691 SRR15101692 SRR15101693 SRR15101694 SRR15101695Downloaded associated SRA runs (done on Mox using a build node):
time \ /gscratch/srlab/programs/sratoolkit.2.11.3-centos_linux64/bin/prefetch \ --option-file /gscratch/srlab/sam/data/NCBI-BioProject-PRJNA744403-coral_metagenomics/SraAccList-PRJNA744403.txtNOTE: This puts all files in this directory:
/gscratch/srlab/data/ncbi/sraTook a bit over 2hrs to download all the files:
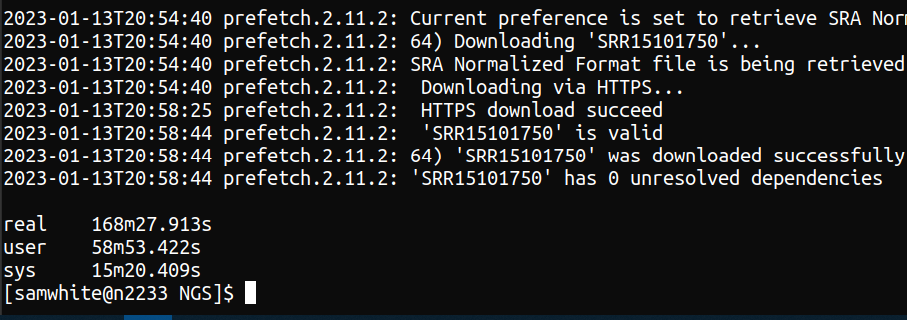
Screenshot showing execution time of 168mins 27.9 seconds to download SRA files for BioProject PRJNA744403 Converted
.srafiles to FastQ (done on Mox using a build node):for file in *.sra; do /gscratch/srlab/programs/sratoolkit.2.11.3-centos_linux64/bin/fasterq-dump "${file}"; doneMoved FastQ files to species-specific directories and library-specific (i.e. BS-seq or RNA-seq) directories.
gzip-ed all FastQ files. This is needed for significant space savings, but also needed for some potential downstream pipelines we’re considering running.Generated MD5 checksums for all files.
Had to run QC/trimming SLURM script for coral SRA data (BioProject PRJNA74403) a couple of times. Seemed like fastp was trimming data (i.e. output reports indicate average read length after filtering is ~140bp, but the resulting graphs of read lengths after trimming still show 150bp…). Additionally, the first ~20bp looked rough, so decided to add a hard 20bp trim from 5’ end of all reads…
FastQC, fastp, and MultiQC were run on Mox.
SBATCH script (GitHub):
#!/bin/bash
## Job Name
#SBATCH --job-name=20230119-coral-fastqc-fastp-multiqc-PRJNA744403
## Allocation Definition
#SBATCH --account=srlab
#SBATCH --partition=srlab
## Resources
## Nodes
#SBATCH --nodes=1
## Walltime (days-hours:minutes:seconds format)
#SBATCH --time=5-00:00:00
## Memory per node
#SBATCH --mem=500G
##turn on e-mail notification
#SBATCH --mail-type=ALL
#SBATCH --mail-user=samwhite@uw.edu
## Specify the working directory for this job
#SBATCH --chdir=/gscratch/scrubbed/samwhite/outputs/20230119-coral-fastqc-fastp-multiqc-PRJNA744403
### FastQC and fastp trimming coral metagenome SRA BioProject PRJNA744403 sequencing data.
### fastp expects input FastQ files to be in format: *_R[12].fastq.gz
###################################################################################
# These variables need to be set by user
## Assign Variables
# Set FastQ filename patterns
fastq_pattern='*.fastq.gz'
R1_fastq_pattern='*_1*.fastq.gz'
R2_fastq_pattern='*_2*.fastq.gz'
# Set species array
species_array=(P_grandis P_meandrina)
# Set number of CPUs to use
threads=40
# Set working directory
working_dir=$(pwd)
# Input/output files
trimmed_checksums=trimmed_fastq_checksums.md5
fastq_checksums=input_fastq_checksums.md5
# Data directories
bsseq_dir="BS-seq"
rnaseq_dir="RNA-seq"
## Inititalize arrays
raw_fastqs_array=()
R1_names_array=()
R2_names_array=()
# Paths to programs
fastp=/gscratch/srlab/programs/fastp.0.23.1
fastqc=/gscratch/srlab/programs/fastqc_v0.11.9/fastqc
multiqc=/gscratch/srlab/programs/anaconda3/bin/multiqc
# Programs associative array
declare -A programs_array
programs_array=(
[fastqc]="${fastqc}"
[fastp]="${fastp}" \
[multiqc]="${multiqc}"
)
###################################################################################
# Exit script if any command fails
set -e
# Load Python Mox module for Python module availability
module load intel-python3_2017
# Capture date
timestamp=$(date +%Y%m%d)
# Sync raw FastQ files to working directory
# Uses "P_[gm]*" to get only files from
# P_grandis and P_meandrina directories
echo ""
echo "Transferring files via rsync..."
rsync --archive \
--verbose \
--progress \
--files-from=<(find ../../data/P_[gm]* -name "*.fastq.gz") \
../../ .
echo ""
echo "File transfer complete."
echo ""
for species in "${species_array[@]}"
do
for library in "${bsseq_dir}" "${rnaseq_dir}"
do
## Re-inititalize arrays
fastq_array_R1=()
fastq_array_R2=()
raw_fastqs_array=()
R1_names_array=()
R2_names_array=()
trimmed_fastq_array=()
# Change to bisulfite data directory
cd "${working_dir}/data/${species}/${library}"
echo ""
echo "Moving to ${working_dir}/data/${species}/${library}"
echo ""
# Create trimmed subdirectory
mkdir --parents trimmed
### Run FastQC ###
### NOTE: Do NOT quote raw_fastqc_list
# Set FastQC output directory
output_dir=$(pwd)
# Create array of raw FastQs
raw_fastqs_array=("${fastq_pattern}")
# Pass array contents to new variable as space-delimited list
raw_fastqc_list=$(echo "${raw_fastqs_array[*]}")
echo "Beginning FastQC on raw reads..."
echo ""
# Run FastQC
${programs_array[fastqc]} \
--threads ${threads} \
--outdir ${output_dir} \
${raw_fastqc_list}
echo ""
echo "FastQC on raw reads complete!"
echo ""
### END FASTQC ###
# Create arrays of fastq R1 files and sample names
# Do NOT quote R1_fastq_pattern variable
for fastq in ${R1_fastq_pattern}
do
fastq_array_R1+=("${fastq}")
# Use parameter substitution to remove all text up to and including last "." from
# right side of string.
R1_names_array+=("${fastq%%.*}")
done
# Create array of fastq R2 files
# Do NOT quote R2_fastq_pattern variable
for fastq in ${R2_fastq_pattern}
do
fastq_array_R2+=("${fastq}")
# Use parameter substitution to remove all text up to and including last "." from
# right side of string.
R2_names_array+=("${fastq%%.*}")
done
# Create MD5 checksums for raw FastQs
for fastq in ${fastq_pattern}
do
echo "Generating checksum for ${fastq}"
md5sum "${fastq}" | tee --append ${fastq_checksums}
echo ""
done
### RUN MULTIQC ###
echo "Beginning MultiQC..."
echo ""
${programs_array[multiqc]} .
echo ""
echo "MultiQC complete."
echo ""
### END MULTIQC ###
### RUN FASTP ###
# Run fastp on files
# Adds JSON report output for downstream usage by MultiQC
echo "Beginning fastp trimming."
echo ""
for index in "${!fastq_array_R1[@]}"
do
R1_sample_name="${R1_names_array[index]}"
R2_sample_name="${R2_names_array[index]}"
${fastp} \
--in1 ${fastq_array_R1[index]} \
--in2 ${fastq_array_R2[index]} \
--detect_adapter_for_pe \
--trim_poly_g \
--trim_front1 20 \
--trim_front2 20 \
--thread ${threads} \
--html trimmed/"SRA-${R1_sample_name%_*}.${species}.fastp-trim.${timestamp}".report.html \
--json trimmed/"SRA-${R1_sample_name%_*}.${species}.fastp-trim.${timestamp}".report.json \
--out1 trimmed/"SRA-${R1_sample_name}.${species}.fastp-trim.${timestamp}".fq.gz \
--out2 trimmed/"SRA-${R2_sample_name}.${species}.fastp-trim.${timestamp}".fq.gz
### END FASTP ###
# Generate md5 checksums for newly trimmed files
{
md5sum trimmed/"SRA-${R1_sample_name}.${species}.fastp-trim.${timestamp}".fq.gz
md5sum trimmed/"SRA-${R2_sample_name}.${species}.fastp-trim.${timestamp}".fq.gz
} >> trimmed/"${trimmed_checksums}"
done
### RUN FASTQC ###
### NOTE: Do NOT quote ${trimmed_fastqc_list}
# Change to trimmed directory
cd trimmed
# Set FastQC output directory
output_dir=$(pwd)
# Create array of trimmed FastQs
trimmed_fastq_array=(*fastp-trim*.fq.gz)
# Pass array contents to new variable as space-delimited list
trimmed_fastqc_list=$(echo "${trimmed_fastq_array[*]}")
# Run FastQC
echo "Beginning FastQC on trimmed reads..."
echo ""
${programs_array[fastqc]} \
--threads ${threads} \
--outdir ${output_dir} \
${trimmed_fastqc_list}
echo ""
echo "FastQC on trimmed reads complete!"
echo ""
### END FASTQC ###
### RUN MULTIQC ###
echo "Beginning MultiQC..."
echo ""
${programs_array[multiqc]} .
echo ""
echo "MultiQC complete."
echo ""
### END MULTIQC ###
done
done
# Return to top directory
cd "${working_dir}"
####################################################################
# Capture program options
if [[ "${#programs_array[@]}" -gt 0 ]]; then
echo "Logging program options..."
for program in "${!programs_array[@]}"
do
{
echo "Program options for ${program}: "
echo ""
# Handle samtools help menus
if [[ "${program}" == "samtools_index" ]] \
|| [[ "${program}" == "samtools_sort" ]] \
|| [[ "${program}" == "samtools_view" ]]
then
${programs_array[$program]}
# Handle DIAMOND BLAST menu
elif [[ "${program}" == "diamond" ]]; then
${programs_array[$program]} help
# Handle NCBI BLASTx menu
elif [[ "${program}" == "blastx" ]]; then
${programs_array[$program]} -help
fi
${programs_array[$program]} -h
echo ""
echo ""
echo "----------------------------------------------"
echo ""
echo ""
} &>> program_options.log || true
# If MultiQC is in programs_array, copy the config file to this directory.
if [[ "${program}" == "multiqc" ]]; then
cp --preserve ~/.multiqc_config.yaml multiqc_config.yaml
fi
done
fi
# Document programs in PATH (primarily for program version ID)
{
date
echo ""
echo "System PATH for $SLURM_JOB_ID"
echo ""
printf "%0.s-" {1..10}
echo "${PATH}" | tr : \\n
} >> system_path.logRESULTS
Runtime was ~6.25hrs:

Due to the large number of files, along with attempts to keep things semi-organized, please refer to the directory-tree first.
To cut right to the chase, the following four files can be downloaded and then used with wget to download all of the described FastQ files.
P.grandis BS-seq raw FastQs
P.grandis BS-seq trimmed FastQs
P.grandis RNA-seq raw FastQs
P.grandis RNA-seq trimmed FastQs
P.meandrina BS-seq raw FastQs
P.meandrina BS-seq trimmed FastQs
P.meandrina RNA-seq raw FastQs
P.meandrina RNA-seq trimmed FastQs
Example of how to use these files:
wget --input-file=pgrandis.bs-seq.raw.download.list.txt
This will download all of the P.grandis BS-seq raw FastQ files and an MD5 checksum file for verifying data after transfer.
Output folder:
20230119-coral-fastqc-fastp-multiqc-PRJNA744403/
Directory tree (text)
NCBI BioProject PRJNA744403 Metadata (CSV)
P.grandis BS-seq raw reads
MultiQCreport (HTML):P.grandis BS-seq trimmed reads
MultiQCreport (HTML):P.grandis RNA-seq raw reads
MultiQCreport (HTML):P.grandis RNA-seq trimmed reads
MultiQCreport (HTML):P.meandrina BS-seq raw reads
MultiQCreport (HTML):P.meandrina BS-seq trimmed reads
MultiQCreport (HTML):P.meandrina RNA-seq raw reads
MultiQCreport (HTML):P.meandrina RNA-seq trimmed reads
MultiQCreport (HTML):