Followed JGI “Bacterial genomic DNA isolation using CTAB” protocol(Word doc) with the following notes/changes.
Two, 1L cultures were transferred to 4 total jars (~500mL in each jar) for centrifugation. Jars had been briefly washed with 95% EtOH prior to use.
Cells were pelleted in Sorval T21 using a bucket rotor for 30mins., 4200RPM, 25C.
Supe was removed and all four pellets were resuspended with a total of 10mL of TE.
OD600 = ~2.1, so added an additional 10mL of TE.
OD600 = ~1.2, so proceeded with procedure.
Due to volume of the prep, the sample was split into two, 50mL centrifuge tubes (washed with 95% EtOH prior to use) prior to the addition of 5M NaCl.
Disaster strikes! Turns out the tubes being used were not resistant to chloroform. This was realized after spinning at 18,000RPM, 25C 10mins in a Sorval SL-50T rotor in the Sorval T21 centrifuge.
However…
I recovered the aqueous phase anyway, as it was still contained in the tube and had no contact with the rotor surface(s). Due to a subsequent phenol:chloroform:IAA step, I split the aqueous phase into 24 x 1.5mL snap cap tubes and proceeded according to protocol. The final DNA pellet, prior to resuspension were combined from all the tubes into a single 1.5mL snap cap tube in a final volume of 400uL of TE.
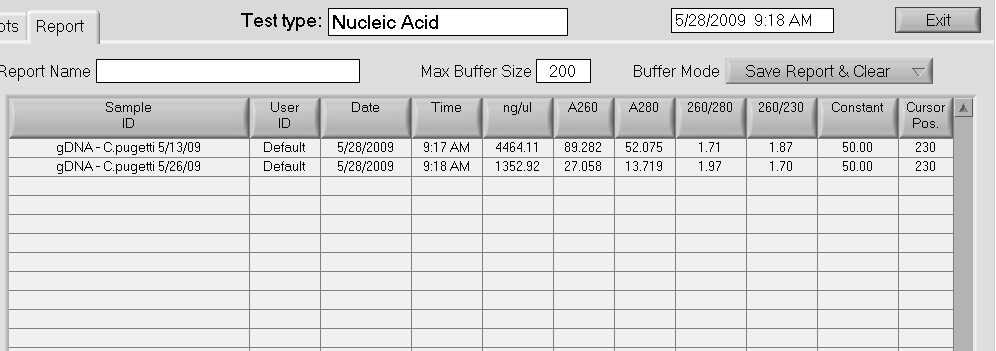
Results: DNA from today looks good with excellent yield. DNA from 20090513 doesn’t look as nice AND the concentration doesn’t jive with the QC gel that was run on 20090513. Will run out on gel according to JGI protocol to evaluate quality further.