Set up PCR on recent Sepia cDNA and DNased RNA samples using both S. officianalis_rhodopsin_F, R primers & Sep_op_F2, R2 primers. After yesterday’s intirguing PCR results, we need to confirm that the DNased RNA is free of contaminating gDNA. Additionally, we want to excise bands from the cDNA samples for sequencing. PCR set up is here. RNA was prepped as though making cDNA; diluted 0.2ug of DNased RNA in a final volume of 25uL. Used 1uL of this for PCRs.
Rhodopsin primers require 3mM Mg2+ in the PCR rxn and the opsin primers require 2mM Mg2+ in the PCR rxn. 25mM Mg2+ was added as required and is shown on the PCR set up link above.
Results: n00b alert! The gel below is a repeat of yesterday’s PCR, except with DNased RNA samples added to verify no gDNA carryover. However, you’ll notice that the PCR results on the cDNA don’t look identical to yesterday’s PCR. Ugh.
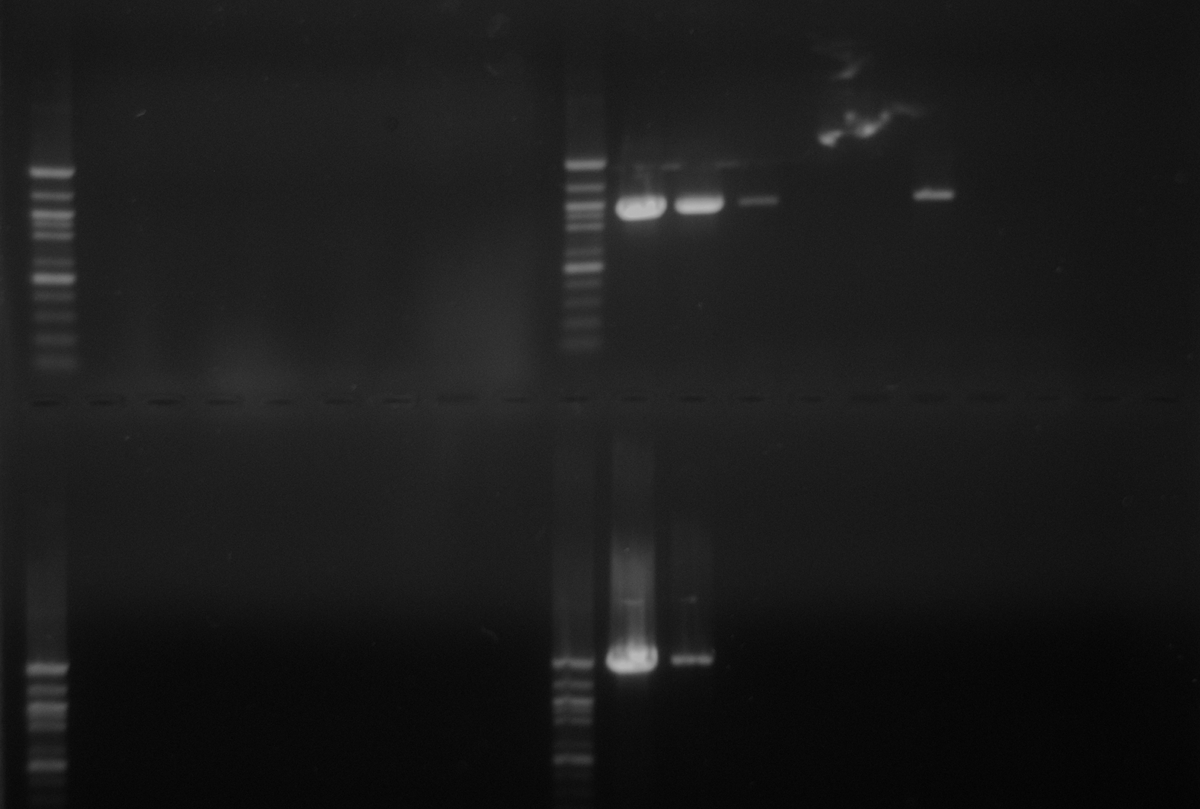
Gel Loading (from left to right):
Opsin Primers (top half of gel), Rhodopsin Primers (same loading order, bottom half of gel)
RNA samples (left sides of gel), cDNA samples (right sides of gel)
1 - 100bp ladder
2 - retina
3 - fin
4 - 4th arm
5 - dorsal mantle center
6 - dorsal mantle side
7 - ventral mantle center
8 - ventral mantle side
9 - H2O
Loading is repeated in the above order for the cDNA, which is the right halves of the gel.
Well, the good news is that no signal is seen in the DNased RNA samples, thus confirming that there is no detectable gDNA carryover.
Opsin Primers
We see a band in the retina, fin & ventral mantle center samples as we did yesterday. However, we see a band in the 4th arm sample, which was not present yesterday. We also do NOT see a band in the ventral mantle side sample like we did yesterday. Negative controls are negative.
Rhodopsin Primers
We see a band in the retina and fin samples as we did yesterday. However, we no longer see the dual bands in the ventral mantle center as were seen yesterday. Negative controls are negative.
Bands were excised from the gel, placed in 1.5mL snap cap tubes and stored in the -20C in Sam’s “Purified Inserts” box.
Because of the lack of repeated results, I will repeat this PCR for a third time, with just the cDNA again, to see if I can duplicate one set of results…