Total and mRNA aliquots (~5ng/uL) were run on the Agilent Bioanalyzer Pico RNA chips.
Results:
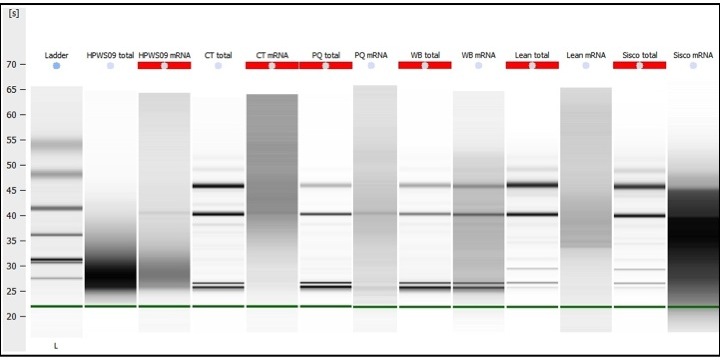
The gel below shows the comparison/results of total RNA and subsequent mRNA isolations. The gel indicates the following:
The HPWS09 total RNA (Herring) is totally degraded, but shows the expected profile in the mRNA prep. It would be extremely interesting to see if the degradation has any effect on sequencing, as the mRNA will get fragmented any way in the next step of library construction.
mRNA isolations worked for all samples. Although one might be inclined to say that mRNA isolation did NOT work for the WB sample, one has to take in to consideration that the gel software adjusts the gel contrast to enhance low signals. That’s why all the mRNA samples exhibit a dark background. mRNA generates a broad, relatively weak signal when compared to a total RNA sample. So, the software attempts to boost the low signal for display purposes. Thus, if we were to decrease this signal boosting (or contrast) for the WB mRNA so that the background color matched the WB total RNA background color (white), the rRNA bands visible in the WB mRNA sample would fade to a point where they would not be visible. See the electropherogram overlay (below the gel) for a more visual comparison of this concept.
Electropherogram Overlays of WB total RNA and WB rRNA
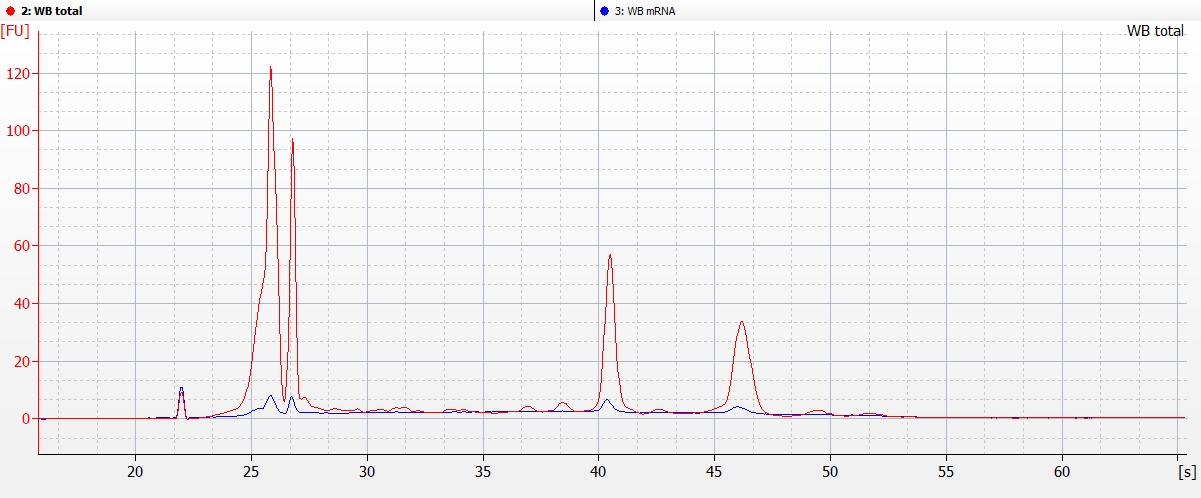
The WB total RNA is the red graph which shows extremely high levels of rRNA (as expected). After subsequent mRNA isolation (the blue graph), the rRNA is virtually gone and no longer comprises a significant portion of the sample.