Performed qPCR using pooled cDNA from 20110311. Pooled 2uL from each of the following samples groups: Dg 3hr C, Gill 1hr C, Gill 1hr E, Mantle 3hr C, and Muscle 3hr C. Master mix calcs are here. Plate layout, cycling params, etc can be found in the qPCR Report (see Results). Primers sets run were:
EF1_qPCR_5’,3’ (SR IDs: 309, 310)
Cg_COX1/2_qPCR_F (SR ID: 1192) + Cg_COX1_qPCR_R (SR ID: 1191)- Target = COX1
Cg_COX1/2_qPCR_F (SR ID: 1192) + Cg_COX2_454align1_R (SR ID: 1190) - Target = COX2
Results:
[qPCR Report (PDF)(https://eagle.fish.washington.edu/Arabidopsis/qPCR/Roberts%20Lab_2011-03-15%2011-35-42_CC009827.pdf)
[qPCR Data File (CFX96)(https://eagle.fish.washington.edu/Arabidopsis/qPCR/Roberts%20Lab_2011-03-15%2011-35-42_CC009827.pcrd)
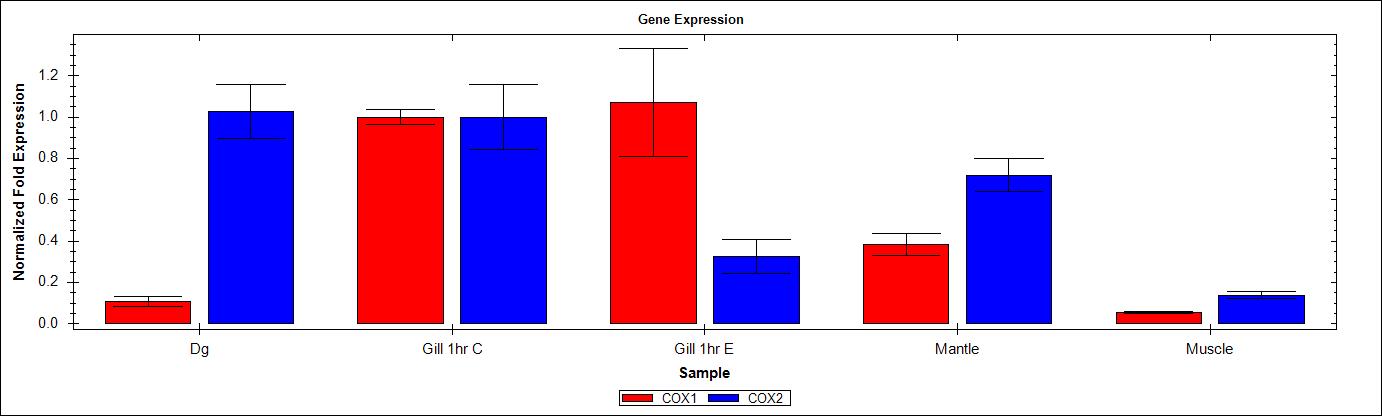
Graphs were generated using the BioRad CFX Manager v2.0 software. Expression was normalized to EF1. Also to note, gene efficiency was assumed as 100% by the software since no standard curve was run on the plate. As such, analysis of this data may not be exact.
It’s clear by examining the graphs that the primers being used to differentiate COX1 and COX2 (since they share a common primer: SRID 1192) are differentially expressed. This indicates that the primer sets are indeed amplifying different targets as hoped. This was the primary intention of this qPCR. However, we also now have an idea of tissue distribution of the two genes, as well as their response to V. vulnificus exposre after 1hr. Next step is to perform this qPCR on all the individuals from this experiment as well as the different tissues.