Due to the recent poor quality gDNA that has been isolated from C.gigas gonad, I decided to do a quick test using TE for DNA pellet resuspension in hopes that old Buffer EB (Qiagen) or old nuclease-free H2O (Promega) are to blame for the apparent, rapid degradation that I’ve experienced.
Isolated gDNA from a C.gigas female gonad sample (EV2 141 go) provided by Mac. Isolated gDNA using DNazol (Molecular Research Center):
Incubated ~25mg of tissue O/N @ RT in 500uL of DNazol + 100ug/mL Proteniase K (2.7uL of 18.5mg/mL Fermentas stock) on rotator.
Added additional 500uL of DNazol and briefly disrupted remaining tissue with a few pipette strokes.
Pelleted debris by spinning 10mins, 10,000g @ RT.
Transferred supe to new tube and repeated Steps 3 & 4 one time.
Added 500uL of 100% EtOH; mixed by inversion.
NOTE: Despite initial appearance of white cloudy appearance after EtOH addition, cloudiness dispersed upon inversion and no visible DNA strands were present
Pelleted DNA by spinning 5000g 5mins @ RT.
Removed supe and washed pellet with 1mL of a 70% DNazol+30% EtOH solution.
Removed supe and washed pellet with 1mL 70% EtOH.
Repeated Step 8 two times.
Discarded supe, quick spun tube to pool residual EtOH. Removed all residual EtOH.
Resuspended in 200uL of TE (pH = 8.0) and incubated at RT for 5mins.
Pelleted insoluble material 12,000g 10mins @ RT.
Transferred supe to clean tube.
Spec’d on NanoDrop1000.
Ran ~500ng on 1.0% agaroase 1x modified TAE gel to evaluate integrity.
Results:
260/280 value looks excellent, but, as always seems to be the case with DNazol/TriReagent, the 260/230 value looks crappy. Will investigate gDNA integrity on agarose gel.
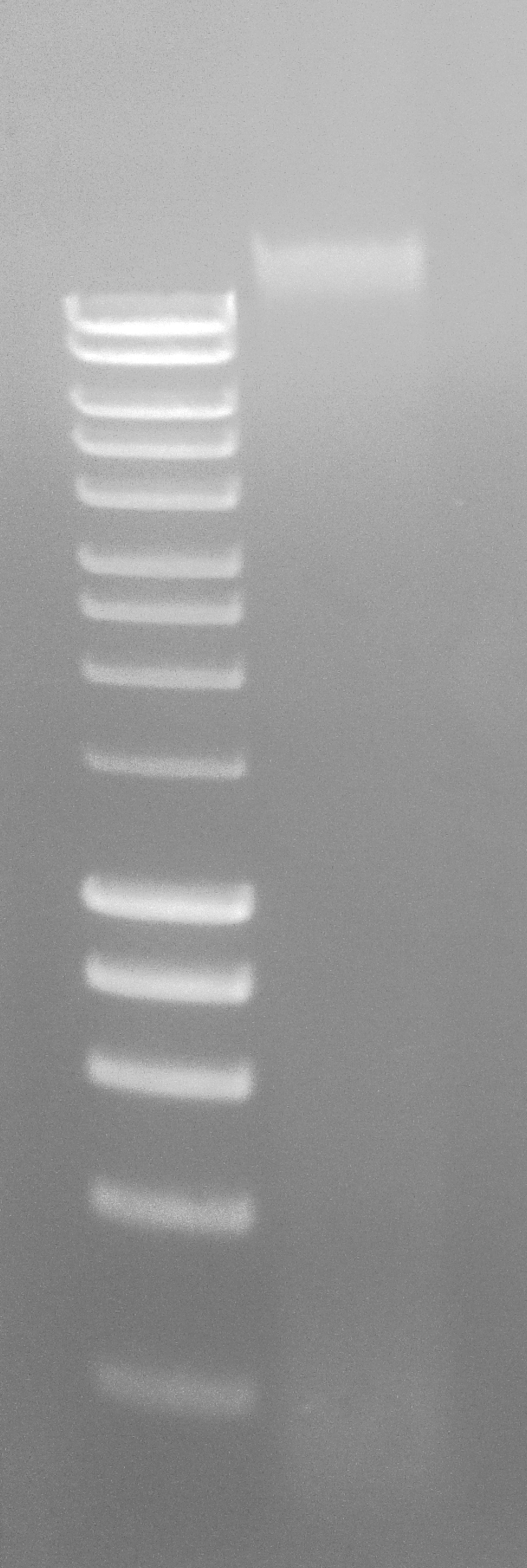
Gel Loading:
Lane 1 - Hyperladder I (Bioline)
Lane 2 - EV2 141 go C.gigas female gonad gDNA
Well, look at that! A nice, clear, high molecular weight band! It looks like my Buffer EB and/or nuclease-free water are is contaminated. Have discarded both. Will re-isolated Claire and Mac’s gDNA.