Zymoresearch support suggested putting the samples through another set of columns to help clean up the apparent phenol carryover that was seen (absorbance peak shifted to 270nm) in the initial isolation of these samples.
Added 500uL of TriReagent to each sample and vortexed. Then, proceeded with the remainder of the protocol (excluding the DNase step). Eluted with 50uL of 0.1% DEPC-treated H2O and spec’d on NanoDrop1000.
Results:
Absolutely horrible!! I can’t even begin to fathom what has happened here. The samples run with the sample kit all worked so well; why did this whole thing have to be jacked up with the actual samples??!!
Well, I’ll do a second elution using 50uL of 0.1%DEPC-treated H2O and spec. Let’s see if that helps….
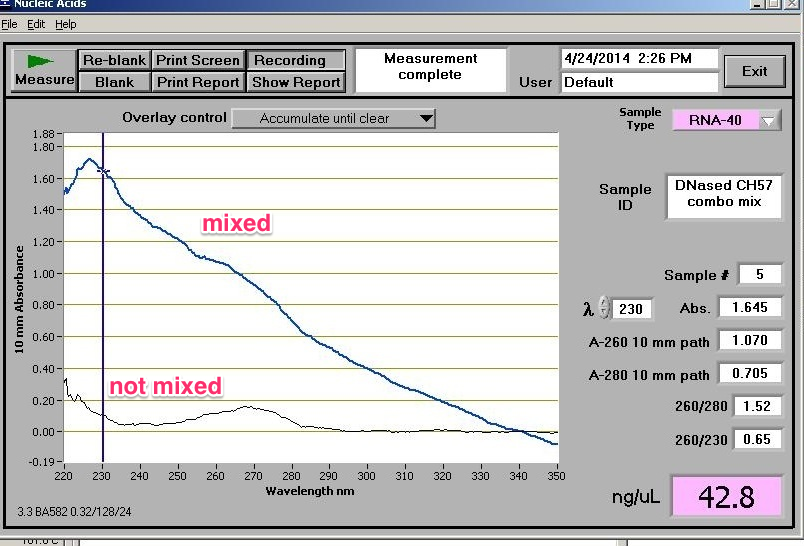
OK, I didn’t even bother spec-ing all the samples because I noticed that the elution tubes had pellets in them! When I mix the tube prior to spec-ing (which is my normal behavior), I get the top absorbance spectra that is virtually useless. When I don’t mix the samples (thus, not disturbing the pellet), I get a more “realistic” spectra, but I can’t tell if I can trust it or not. I have contacted Zymoresearch support for more help with this…
It’s tempting to simply proceed with an EtOH precipitation, but I’m a bit concerned that the pellet in the tubes is resin from the column and that it might still bind some of the RNA. However, I guess the pellet is already in the elution solution, so the RNA should be soluble and, theoretically, not be able to bind to any residual resin…