Trying this sample again(!!), but will now use TE for pellet resuspension to prevent sample degradation. Incubated sample RT on rotator in 500uL of DNazol + 2.7uL of Proteinase K (Fermentas; Stock 18.5mg/mL) for 5hrs. Added additional 500uL of DNazol, mixed gently and followed DNazol manufacturer’s protocol. Performed first pellet was with 70% DNazol/ 30% EtOH solution. Resuspended pellet in 200uL of TE and spec’d on NanoDrop1000.
Results:
Yield is good. 260/280 value is good. 260/230 value is poor. Will run on gel to evaluate integrity.
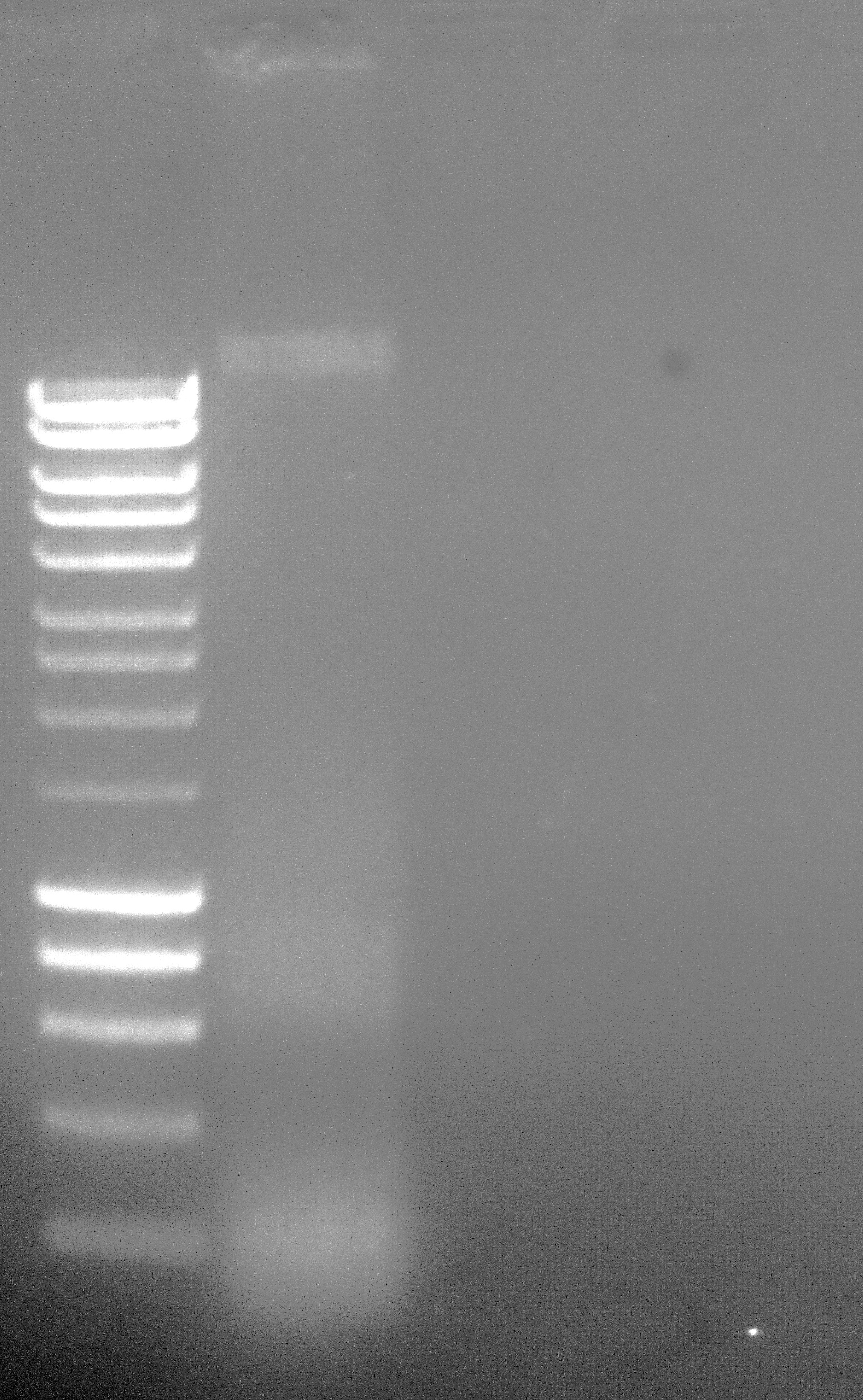
Loaded 10uL (~830ng) on 1.0% agarose 1x modified TAE gel stained with EtBr.
Gel Loading Guide:
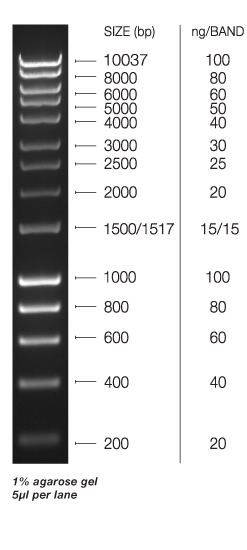
Lane 1 - Hyperladder I (Bioline)
Lane 2 - C.gigas female gonad gDNA (CgF)
Well, this certainly looks much better than previous preparations, in that there is an obvious high molecular weight band present (previously, this had been absent). The low molecular weight bands/smears are possibly RNA carryover and/or degraded DNA. Will discuss with Steven and then, most likely, bring downtown for Illumina sequencing.
UPDATE 20140508: Downtown sequencing facility says there’s only ~800ng of DNA! This is a far cry from the minimum amount needed for sequencing (6ug). Looking at the gel above and comparing sample band intensity to the ladder band intensities suggests that the downtown sequencing facility is correct. I loaded 10uL of DNA on the gel and the intensity of the high molecular weight band is similar to the 400bp band intensity. This corresponds to 40ng of DNA. That means the CgF gDNA band is 40ng/10uL = 4ng/uL. I resuspended the gDNA pellet in 200uL of TE, so 200uL x 4ng/uL = 800ng; exactly what the sequencing facility says they measured…
I’m not entirely sure what is happening here. Until very recently, there were almost never such egregious differences between the NanoDrop measurements and what they were measuring downtown at the sequencing facility. It seems as though they have changed the way they quantify samples (possibly using an Agilent Bioanalyzer instead of the Life Technologies Qubit fluorometer?), but this doesn’t mean their measurements are incorrect. However, I’m starting to suspect that the reason the initial sequencing of this sample was due to an overestimation of the quantity of input DNA (since I believe they were still using the fluorometer back then).
As such, it’s become clear that C.gigas gonad samples seem to yield poor quantities of gDNA, relative to the amount of input material. Additionally, there may be insufficient sample left to generate a useable quantity of gDNA to complete this resequencing effort.