Per Mac’s request, ran a PCR on a set of bisulfite-treated DNA (in her gDNA 2014 box in small -20C):
EV2.16 bisulfite
EV2.20 bisulfite
EV2.22 bisulfite
EV2.24 bisulfite
EV2.28 bisulfite
EV2.29 bisulfite
EV2.32 bisulfite
EV2.33 bisulfite
DNA needed to be diluted. Diluted according to this sheet provided by Mac:
https://eagle.fish.washington.edu/bivalvia/070914bisulfite.pdf
NOTE: EV2.28 didn’t have sufficient DNA left to prepare the dilution according to Mac’s sheet. Instead, the remaining volume ofEv2.28 bisulfite DNA (0.5uL) was diluted in a total volume of 2.5uL to maintain the same dilution ratio.
Master mix calcs are here: 20140828 - PCR Mac Bisulfite Samples
Primers used were:
CgBS_733_26796Seq (SRID: 1598)
CgBS_733_26796R_5’biotin (SRID: 1596)
Cycling params:
- 95C - 10mins
- 94C - 30s
- 56C - 30s
- 72C - 30s
- Repeat steps 2 - 5 44 more times
- 72C - 10mins
Results:
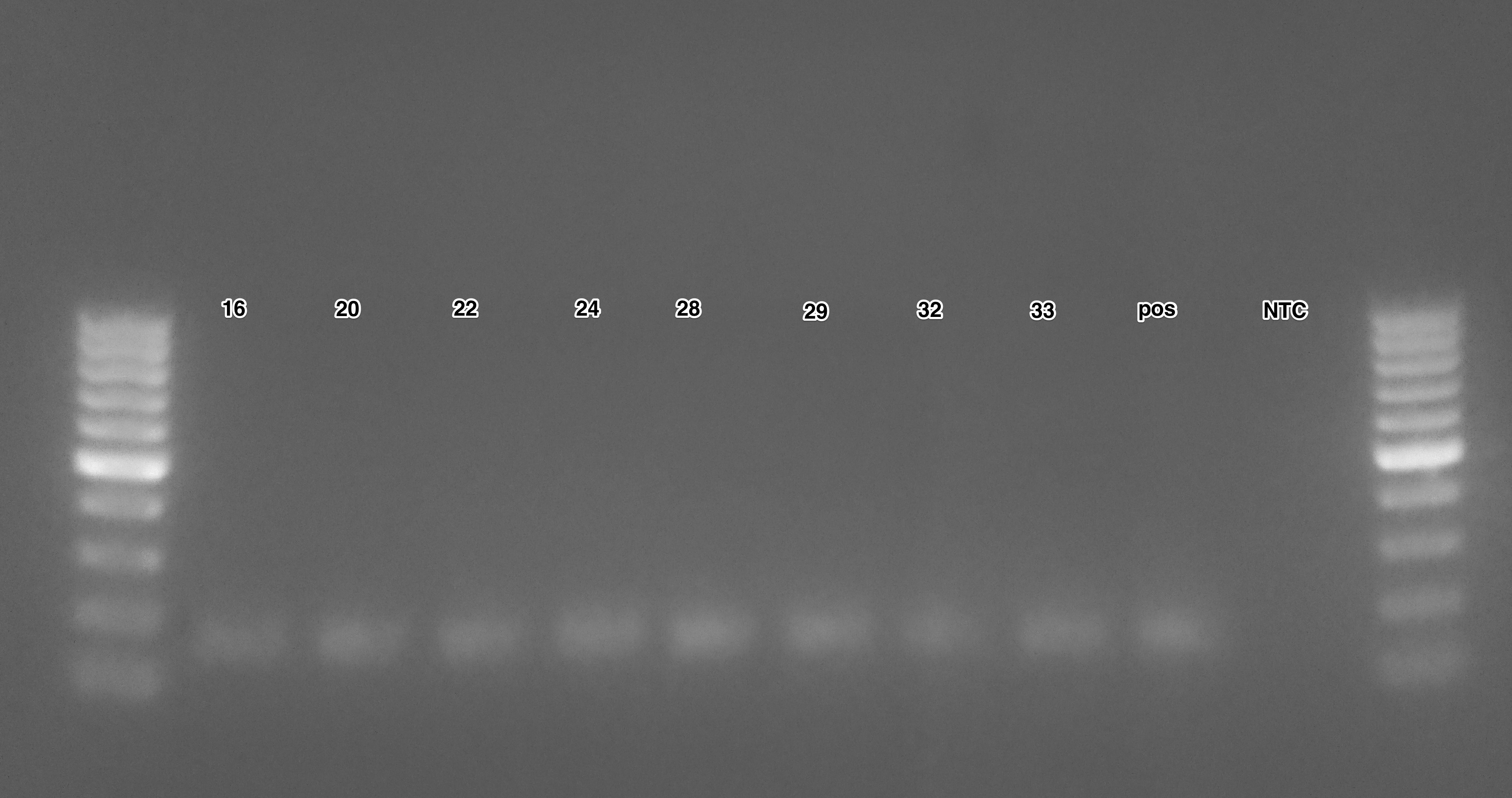
Ladder used is O’GeneRuler 100bp DNA Ladder (ThermoFisher).
According to Mac, the expected band size is ~300bp. However, all samples are running at ~150bp. Mac is confused and does not know what to do.
UPDATE 20140902 - Realized I used the wrong forward primer! Will repeat PCR with correct primer. Wonder if Mac did the same thing…