I was contacted by the sequencing facility at the University of Oregon regarding a sample quality issue with our library. As evidenced by the electropherogram below, there is a great deal of adaptor primer dimer (the peak at 128bp):
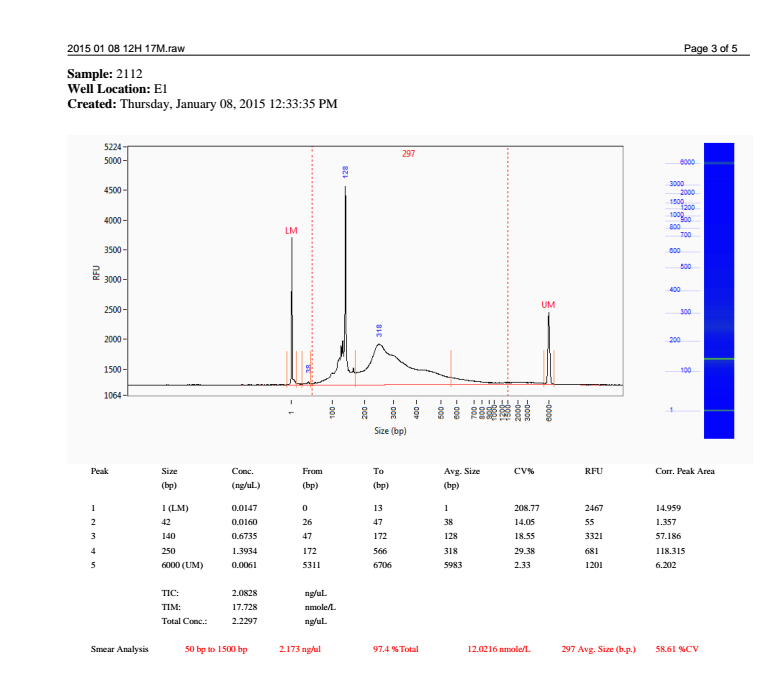 (http://eagle.fish.washington.edu/Arabidopsis/20150120_LSUoilNGSlibraryBioanalyzer.png)
(http://eagle.fish.washington.edu/Arabidopsis/20150120_LSUoilNGSlibraryBioanalyzer.png)
This is a problem because such a high quantity of adaptor sequence will result in the majority of reads coming off the Illumina being just adaptor sequences.
With the remainder of the library sample prepared earlier, I performed the recommended clean up procedure for removing adaptor sequences in the EpiNext Post-Bisulfite DNA Library Preparation Kit – Illumina (Epigentek). Briefly:
Brought sample volume up to 20uL with NanoPure H2O (added 9.99uL)
Added equal volume of MQ Beads
Washed beads 3x w/80% EtOH
Eluted DNA w/12uL Buffer EB (Qiagen)
After clean up, quantified the sample via fluorescence using the Quant-iT DNA BR Kit (Life Technologies/Invitrogen). Used 1uL of the sample and the standards. All standards were run in duplicate and read on a FLx800 plate reader (BioTek).
Results are here: 20150122 - LSU_virginicaMBDlibraryCleanup
Library concentration = 2.46ng/uL
Brought the entire sample up to 20uL with Buffer EB (Qiagen) and a final concentration of 0.1% Tween-20 (required by the sequencing facility).
Sent sample to the University of Oregon to replace our previous submission.