Before proceeding with transcriptomics for this project, we need to assess the integrity of the RNA via Bioanalyzer.
RNA that was previously isolated on 20150508, 20150427, and 20150424 (those notebook entries have been updated to report this consolidation and have a link to this notebook entry) were consolidated into single samples (if there had been multiple isolations of the same sample) and spec’d on the Roberts Lab NanoDrop1000:
(http://eagle.fish.washington.edu/Arabidopsis/20150528_geoduck_histo_RNA_ODs.JPG)
(http://eagle.fish.washington.edu/Arabidopsis/20150528_geoduck_histo_RNA_plots.JPG)
Google Sheet: 20150528_geoduck_histo_RNA_ODs
NOTE: Screwed up consolidation of Geoduck Block 03 sample (added one of the 04 dupes to the tube, so discarded 03).
RNA was stored in Shellfish RNA Box #5.
RNA was submitted to to Jesse Tsai at University of Washington Department of Environmental and Occupational Health Science Functional Genomics Laboratory for running on the Agilent Bioanalyzer 2100, using either the RNA Pico or RNA Nano chips, depending on RNA concentration (Pico for lower concentrations and Nano for higher concentrations - left decision up to Jesse).
Results:
Bioanalzyer 2100 Pico Data File (XAD): SamWhite_Eukaryote Total RNA Pico_2015-05-28_12-50-00.xad Bioanalzyer 2100 Nano Data File (XAD): SamWhite_Eukaryote Total RNA Nano_2015-05-28_13-22-53.xad
Pico Gel Representation
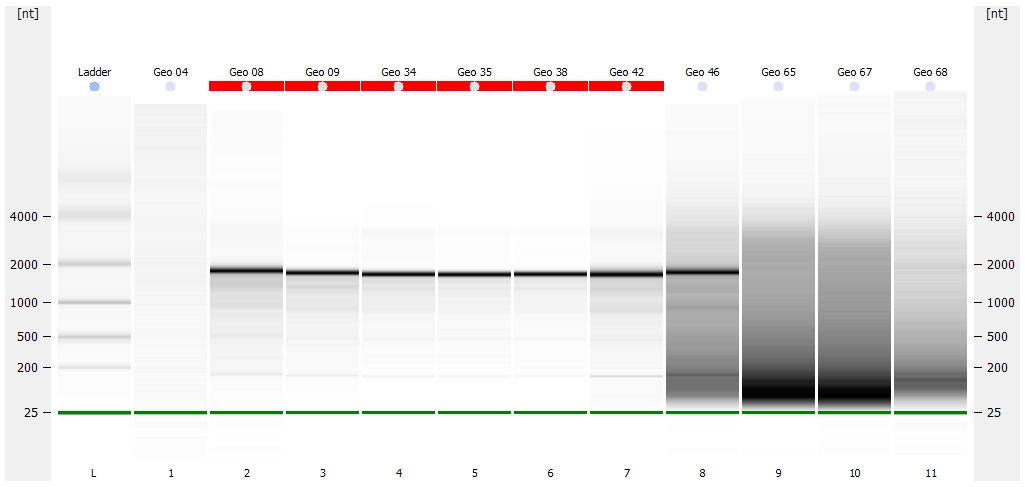 (http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_pico_geoduck_gel.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_pico_geoduck_gel.jpg)
Pico Electropherogram
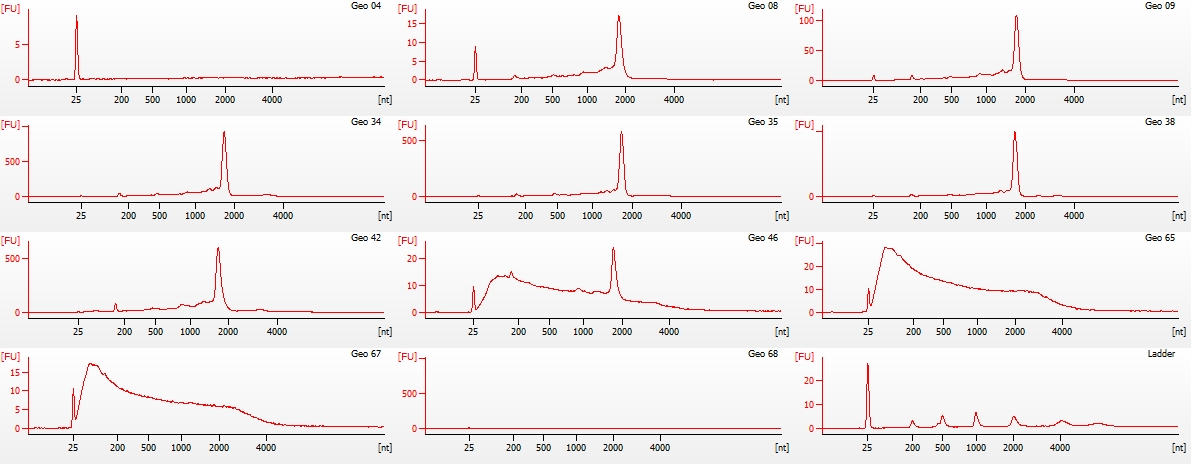 (http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_pico_geoduck_electropherogram.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_pico_geoduck_electropherogram.jpg)
Nano Gel Representation
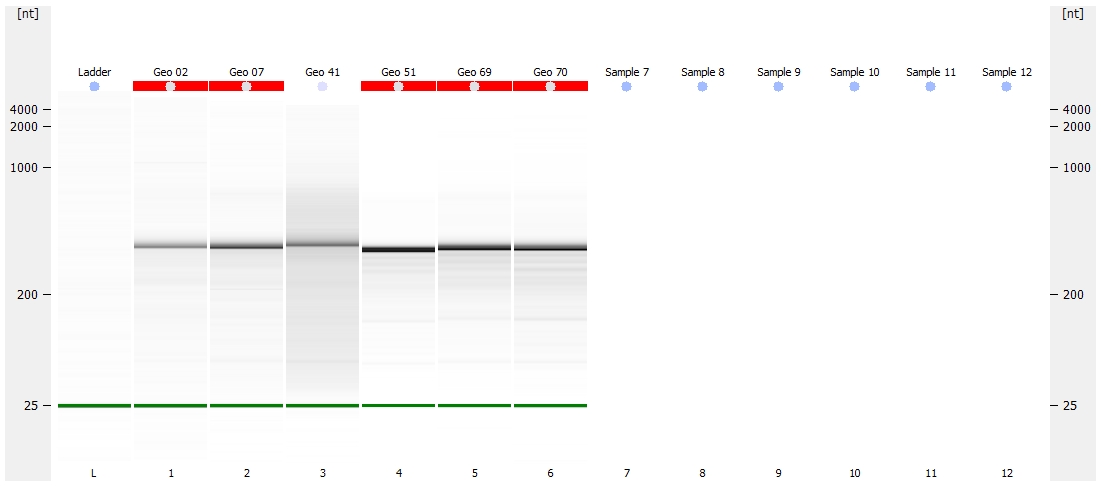 (http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_nano_geoduck_gel.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_nano_geoduck_gel.jpg)
Nano Electropherogram
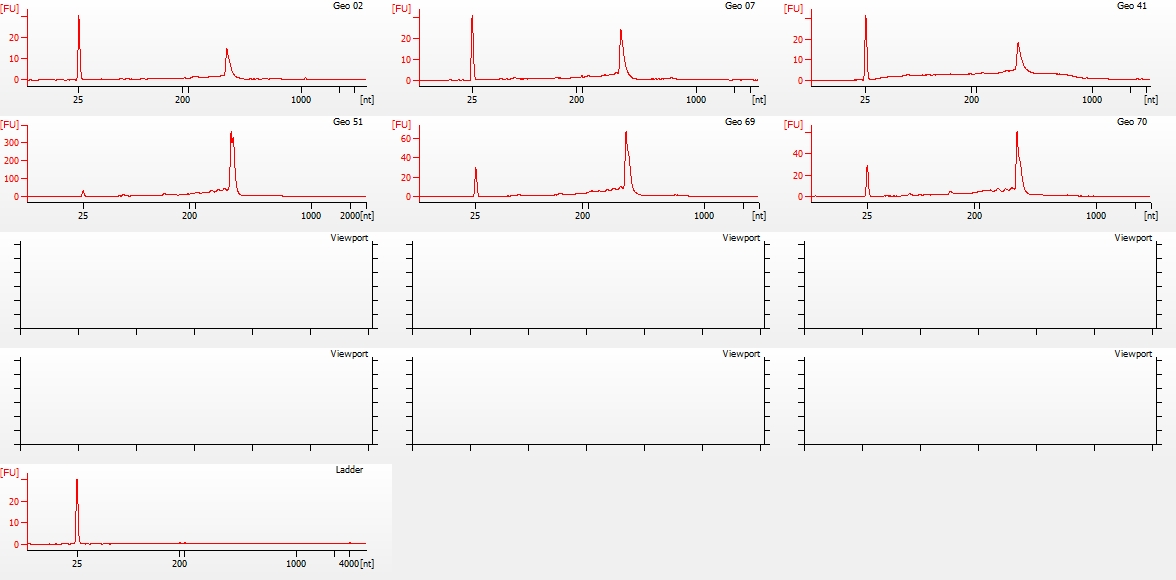 (http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_nano_geoduck_electropherogram.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/Bioanalyzer%20Data/20150528_bioanalyzer_RNA_nano_geoduck_electropherogram.jpg)
Jesse alerted me to the fact that they did not have any ladder to use on the Nano chip, as someone had used the remainder, but failed to order more. I OK’d him to go ahead with the Nano chip despite lacking ladder, as we primarily needed to assess RNA integrity.
Bad Samples:
Geo 04 - No RNA detected
Geo 65, 67, 68 - These three samples show complete degradation of the RNA (i.e. no ribosomal band present, significant smearing on the gel representation).
All other samples look solid. Will discuss with Steven and Brent on how they want to proceed.
Full list of samples for this project (including the Block 03 sample not included in this analysis; see above). Grace’s notebook(previouslly avaialable in genefish.wikispaces.com/Grace%27s+Notebook) will have details on what the numbering indicates (e.g. developmental stage).
block 02
block 03 (no RNA)
block 04 (no RNA)
block 07
block 08
block 09
block 34
block 35
block 38
block 41
block 42
block 46
block 51
block 65 (degraded RNA)
block 67 (degraded RNA)
block 68 (degraded RNA)
block 69
block 70