Ran a PCR to obtain luciferase DNA for sequencing.
Used sea pen gDNA extracted by Jonathan on 20150527.
Primers:
| SRID | Name |
| 1604 | Rr_46_65F |
| 1603 | Rr_887_868R |
Master mix calcs are here: 20150702_seapen_PCR
Cycling params:
95C - 10mins
95C - 15s
55C - 15s
72C - 1min
Go to Step 2 39 times
Ran samples on 0.8% agarose, low TAE gel stained with EtBr.
Results:
 (https://github.com/sr320/LabDocs/blob/master/protocols/Commercial_Protocols/ThermoFisher_OGeneRuler100bpDNA_ladder.jpg?raw=true)
(https://github.com/sr320/LabDocs/blob/master/protocols/Commercial_Protocols/ThermoFisher_OGeneRuler100bpDNA_ladder.jpg?raw=true)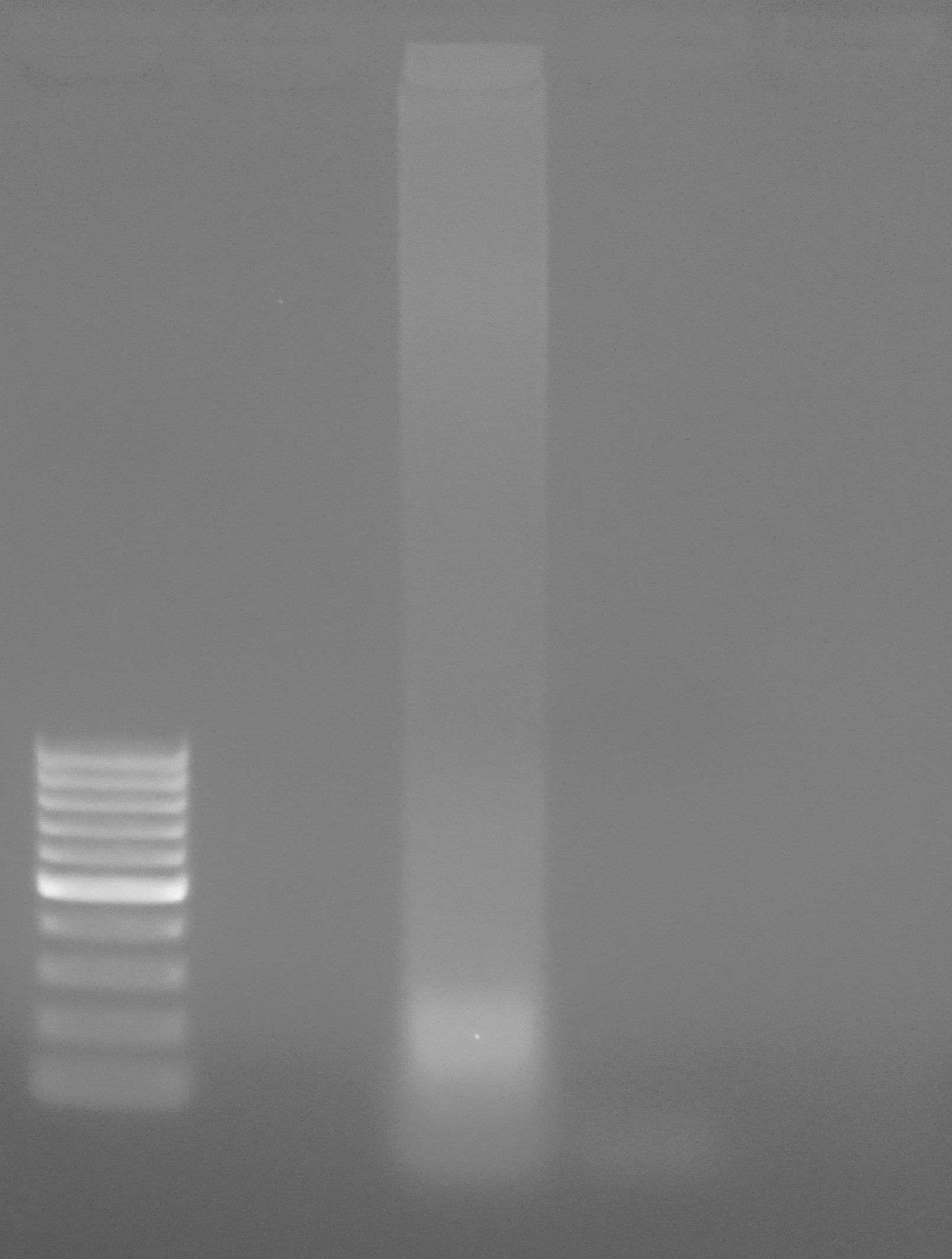
Loading:
Lane 1 - ladder
Lane 2 - empty
Lane 3 - sea pen gDNA
Lane 4 - NTC
PCR did not work. Was expecting a band of ~800bp.
Looks like I may have overloaded the PCR reaction with gDNA. Used 10μL of gDNA.
However, that is quite the smear, suggesting a significant amount of degradation present in the gDNA.
Will re-run this PCR next week with less gDNA (or, cDNA instead) in order to generate a PCR product.