Continuing with the RAD-seq library prep. Following the Meyer Lab 2bRAD protocol. After determining the minimum number of PCR cycles to run to generate a visible, 166bp band on a gel yesterday, ran a full library “prep scale” PCR.
| REAGENT | SINGLE REACTION (μL) | x11 |
| Template | 40 | NA |
| ILL-HT1 (1μM) | 5 | 55 |
| ILL-BC# (1μM) | 5 | NA |
| NanoPure H2O | 5 | 55 |
| dNTPs (1mM) | 20 | 220 |
| ILL-LIB1 (10μM) | 2 | 22 |
| ILL-LIB2 (10μM) | 2 | 22 |
| 5x Q5 Reaction Buffer | 20 | 220 |
| Q5 DNA Polymerase | 1 | 11 |
| TOTAL | 100 | 550 |
Combined the following for PCR reactions:
55μL PCR master mix
40μL ligation mix
5μL of ILL-BC# (1μM) – The barcode number and the respective sample are listed below.
| SAMPLE | BARCODE | SEQUENCE |
| Oly RAD 02 | 1 | CGTGAT |
| Oly RAD 03 | 2 | ACATCG |
| Oly RAD 04 | 3 | GCCTAA |
| Oly RAD 06 | 4 | TGGTCA |
| Oly RAD 07 | 5 | CACTGT |
| Oly RAD 08 | 6 | ATTGGC |
| Oly RAD 14 | 7 | GATCTG |
| Oly RAD 17 | 8 | TCAAGT |
| Oly RAD 23 | 9 | CTGATC |
| Oly RAD 30 | 10 | AAGCTA |
Cycling was performed on a PTC-200 (MJ Research) with a heated lid:
| STEP | TEMP (C) | TIME (s) |
| Initial Denaturation |
|
|
| 17 cycles |
|
|
After cycling, added 16μL of 6x loading dye to each sample.
Loaded 10μL of ladder on each of the two gels.
Results:
 (https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
(https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
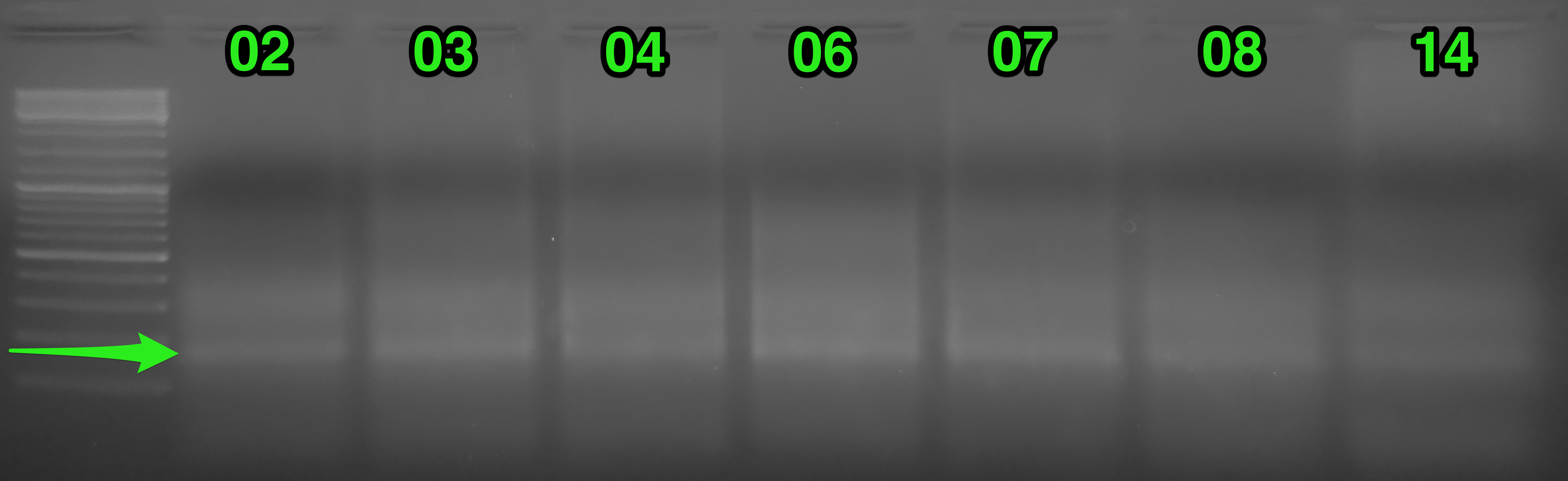 (http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_01.png)
(http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_01.png)
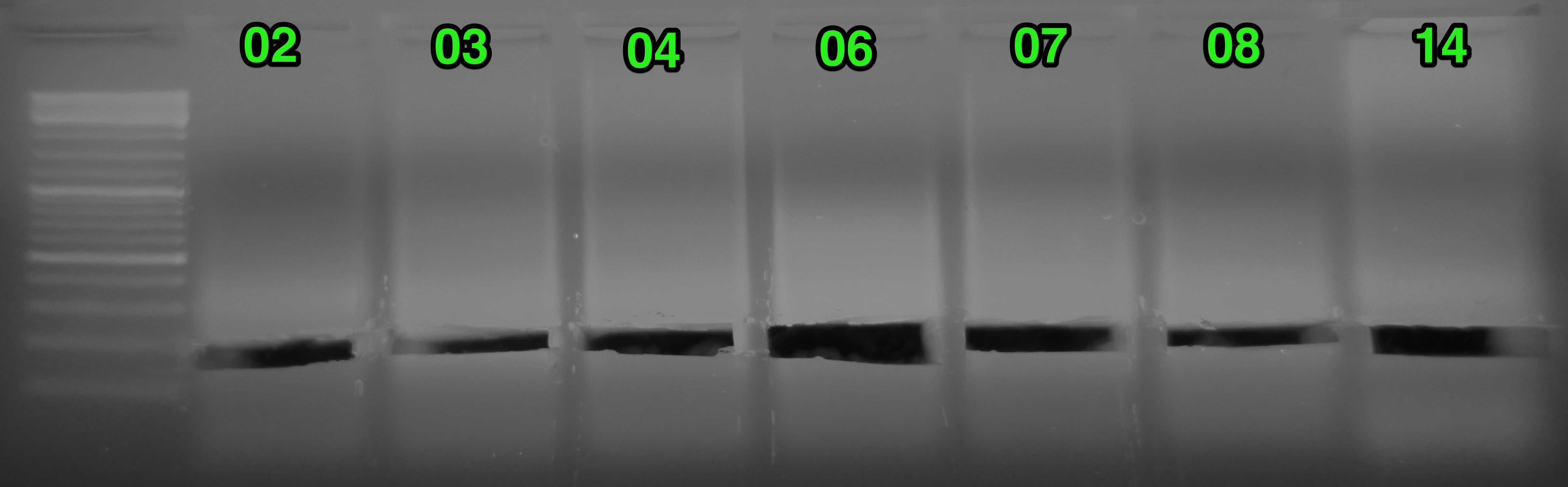 (http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_02.png)
(http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_02.png)
 (http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_03.png)
(http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_03.png)
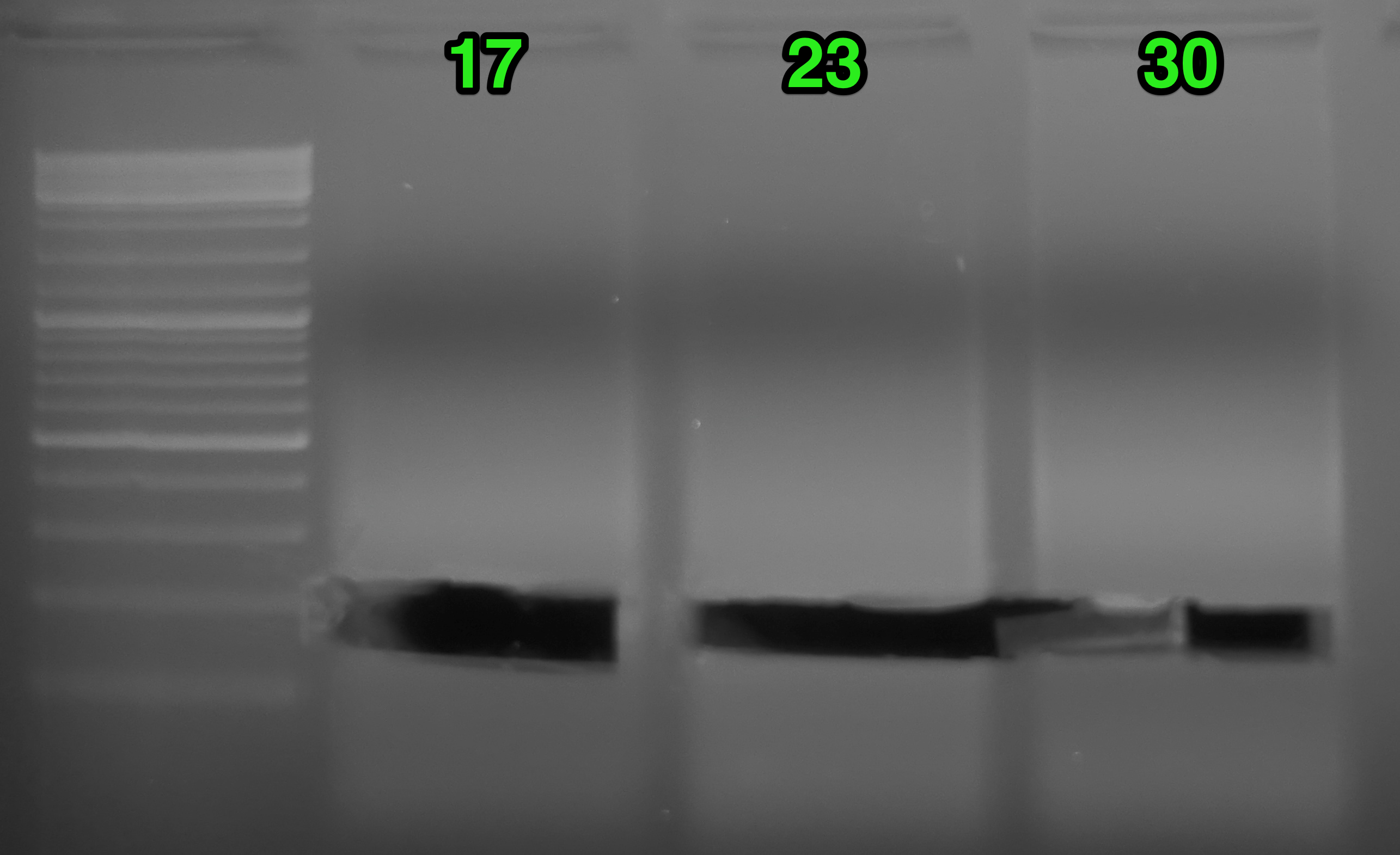 (http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_04.png)
(http://eagle.fish.washington.edu/Arabidopsis/20151113_gel_oly_RAD_prep_PCR_04.png)
Things looked fine. Excised the bands from each sample indicated by the green arrow. Before and after gel images show regions excised. Will purify the bands and quantify library yields.