Have three separate sets of geoduck & olympia oyster gDNA that need to be run on gels before sending to BGI for genome sequencing:
GEODUCK
OLYMPIA OYSTER
Ran 100ng of each sample on a 0.8% agarose 1x modified TAE gel w/EtBr.
Results:
 (https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
(https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
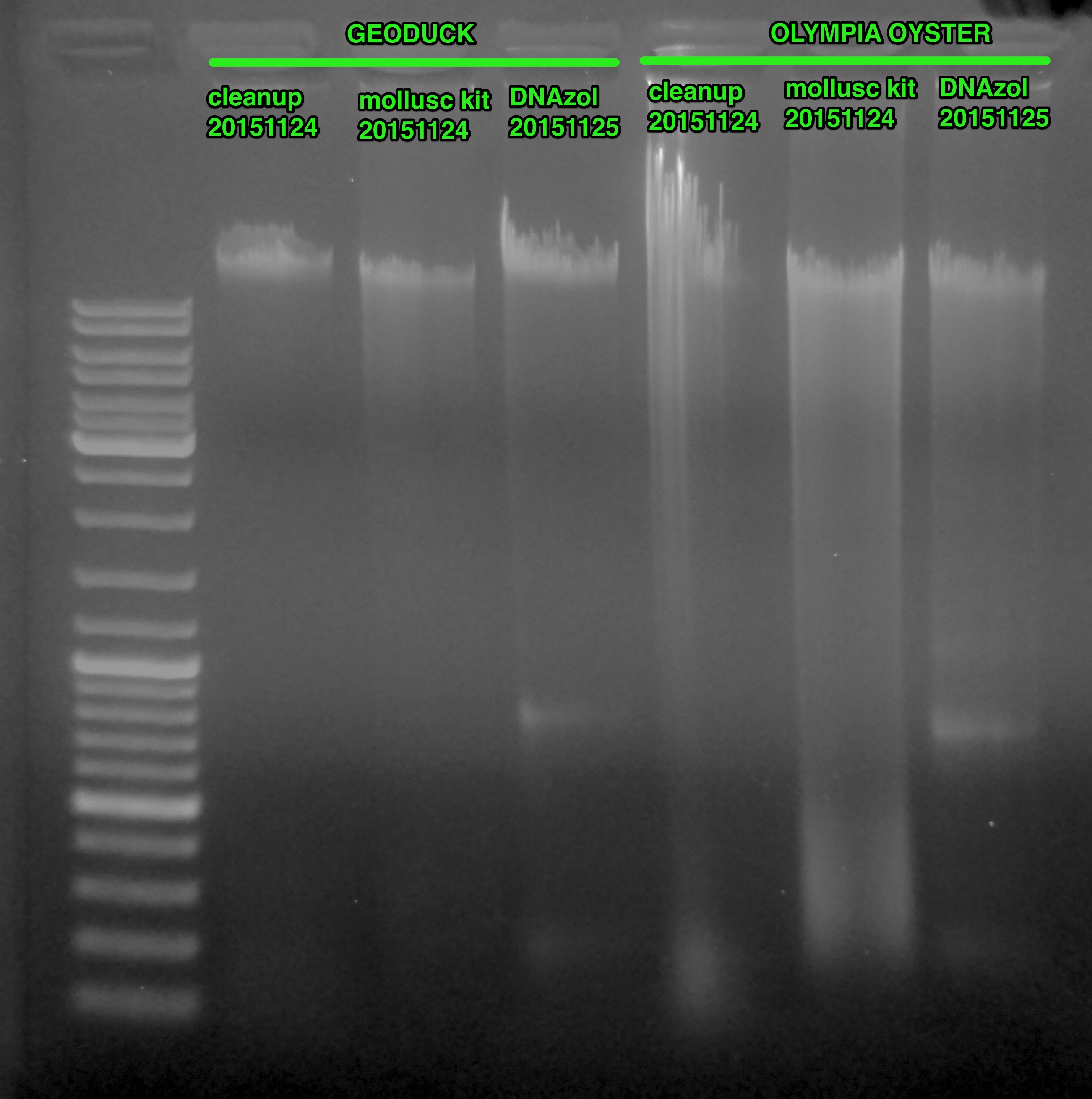 (http://eagle.fish.washington.edu/Arabidopsis/20151125_gel_gDNA_geoduck_oly.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20151125_gel_gDNA_geoduck_oly.jpg)
All the samples from both sets appear to be overloaded. Overloading is generally seen as the streaking seen immediately above each band.
GEODUCK
Overall, the samples look pretty good. Sadly, the worst of the three (due to the most smearing - i.e. degradation) appears to be the DNA extracted using the E.Z.N.A. Mollusc Kit (Omega BioTek).
Also of note are the two bands present in the DNAzol sample. These bands are likely ribosomal RNA because I neglected to perform a RNase treatment during the extraction. Doh!
OLYMPIA OYSTER
None of them are particularly great. Just like the geoduck set, the worst of the three came from the E.Z.N.A Mollusc Kit (Omega BioTek).
Also, just like the geoduck set, there are two bands present in the DNAzol sample. These bands are likely ribosomal RNA because I neglected to perform a RNase treatment during the extraction. Doh!
The phenol-chloroform clean up sample is either jacked up or severely overloaded, based on the crazy streaking that’s present. However, this sample looked similar after the initial extraction on 20151113.
I will send these samples separately (i.e. will not pool them into single samples) to BGI to run QC and, hopefully, add them to the DNA they already have to complete the genome sequencing for these two projects.