The gel I ran earlier today looked real rough, due to the fact that I didn’t bother to equalize loading quantities of samples (I just loaded 1μL of all samples regardless of concentration). So, I’m repeating it using 100ng of DNA from all samples.
Additionally, this gel also includes C.gigas samples that Katie Lotterhos sent to us to see how they look.
Ran a 0.8% agarose, low-TAE gel, stained with ethidium bromide.
Results:  (https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
(https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
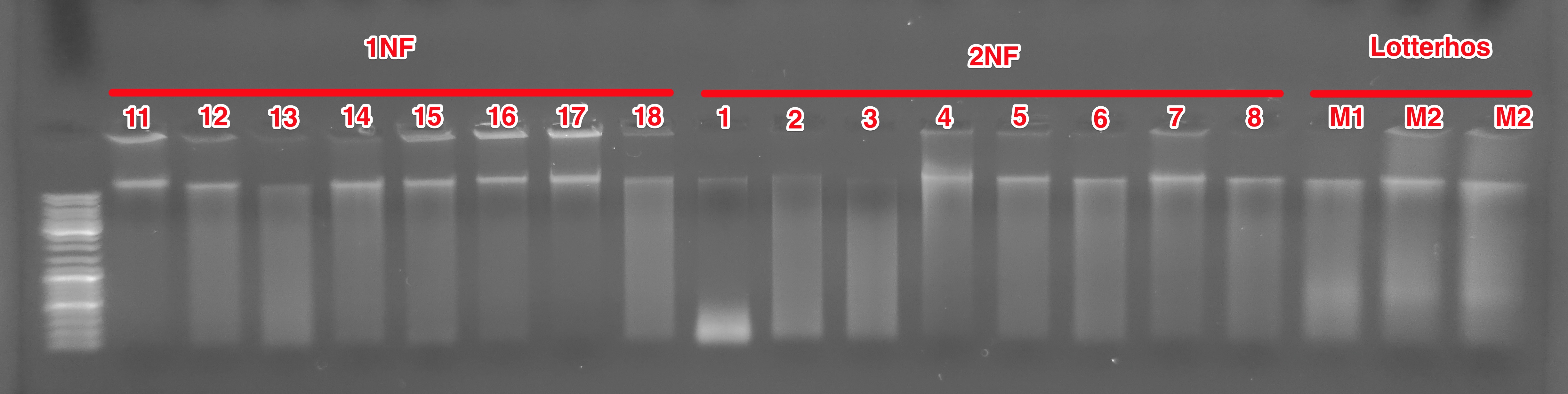 (http://eagle.fish.washington.edu/Arabidopsis/20151217_gel_Oly_gDNA_02.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20151217_gel_Oly_gDNA_02.jpg)
Look at that! The samples look MUCH nicer when they’re not overloaded and uniformly loaded!
Most have a prominent high molecular weight band (the band that’s closes to the top of the ladder, not the DNA visible in the wells). All exhibit smearing, but 2NF1 shows a weird accumulation of low molecular weight DNA.
Katie’s C.gigas samples (M1, M2, M3) look similar to the Olympia oyster gDNA, however her samples appear to have residual RNA in them (the fuzzy band ~500bp).
Will discuss with Steven which samples he wants to use for bisulfite treament and library construction.