Following the guidelines of the [TruSeq DNA Methylation Library Prep Guide (Illumina)(https://github.com/sr320/LabDocs/blob/master/protocols/Commercial_Protocols/Illumina_truseq-dna-methylation-library-prep-guide-15066014-a.pdf), I ran 1μL of each sample on an RNA Pico 6000 chip on the Seeb Lab’s Bioanalyzer 2100 (Agilent) to confirm that bisulfite conversion from earlier today worked.
Results:
Data File 1(Bioanlyzer 2100): 2100 expert_Eukaryote Total RNA Pico_DE72902486_2015-12-18_21-05-04.xad
Data File 1(Bioanlyzer 2100): 2100 expert_Eukaryote Total RNA Pico_DE72902486_2015-12-18_21-42-55.xad
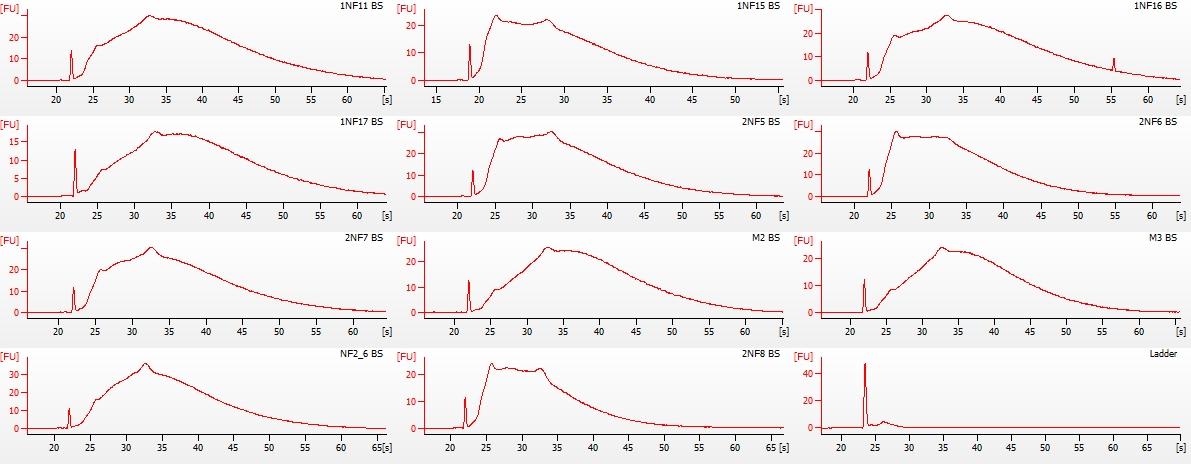 (http://eagle.fish.washington.edu/Arabidopsis/20151218_bioanalyzer_RNApico_oly_bisulfite_01.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20151218_bioanalyzer_RNApico_oly_bisulfite_01.jpg)
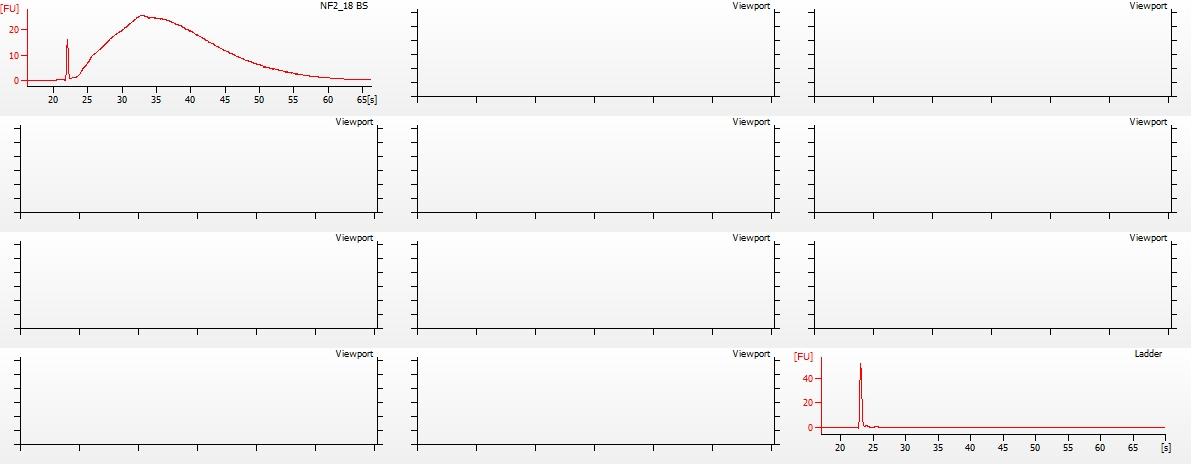 (http://eagle.fish.washington.edu/Arabidopsis/20151218_bioanalyzer_RNApico_oly_bisulfite_02.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20151218_bioanalyzer_RNApico_oly_bisulfite_02.jpg)
Firstly, the ladder failed to produce any peaks. Not sure why this happened. Possibly not denatured? Seems unlikely, but next time I run the Pico assay, I’ll denature the ladder aliquot I use prior to running.
Overall, the samples look as they should (see image from TruSeq DNA Methylation Kit manual below), albeit some are a bit lumpy.
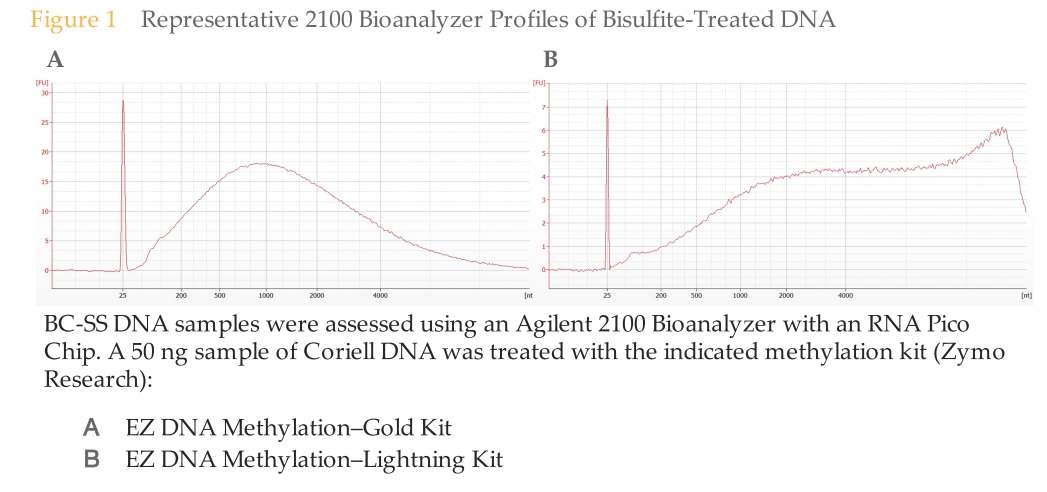 (http://eagle.fish.washington.edu/Arabidopsis/20151218_bioanalyzer_illumina_bisulfite.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20151218_bioanalyzer_illumina_bisulfite.jpg)