In an attempt to improve upon the partial genome assembly we received from BGI, we will be sending DNA to the UW PacBio core facility for additional sequencing.
Isolated DNA from mantle tissue from the same Ostrea lurida individual used for the BGI sequencing efforts. Tissue was collected by Brent & Steven on 20150812 ( dead link - onsnetwork.org/halfshell/2015/08/12/another-day-another-species/).
Used the E.Z.N.A. Mollusc Kit (Omega) to isolate DNA from two separate 50mg pieces of mantle tissue according to the manufacturer’s protocol, with the following changes:
Samples were homogenized with plastic, disposable pestle in 350μL of ML1 Buffer
Incubated homogenate at 60C for 1.5hrs
No optional steps were used
Performed three rounds of 24:1 chloroform:IAA treatment
Eluted each in 50μL of Elution Buffer and pooled into a single sample
Quantified the DNA using the Qubit dsDNA BR Kit (Invitrogen). Used 1μL of DNA sample.
Concentration = 326ng/μL (Quant data is here [Google Sheet]: 20161214_gDNA_Olurida_qubit_quant
Yield is good and we have more than enough (~5μg is required for sequencing) to proceed with sequencing.
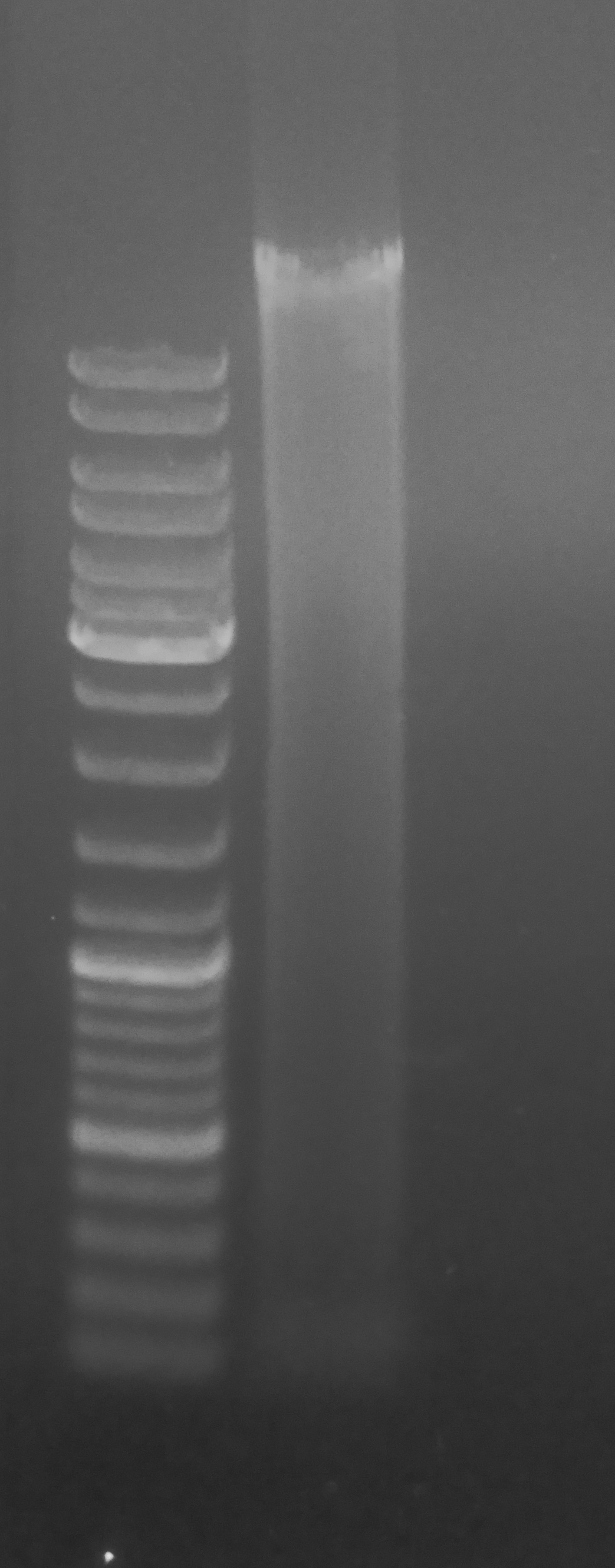
Evaluated gDNA quality (i.e. integrity) by running ~500ng (1.5μL) of sample on 0.8% agarose, low-TAE gel stained with ethidium bromide.
Used 5μL of O’GeneRuler DNA Ladder Mix (ThermoFisher).
Results:
 (https://github.com/sr320/LabDocs/blob/9073dc7caf2dcf40e7739fc7ce9d922b28468dc3/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg?raw=true)
(https://github.com/sr320/LabDocs/blob/9073dc7caf2dcf40e7739fc7ce9d922b28468dc3/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg?raw=true)
 (http://eagle.fish.washington.edu/Arabidopsis/20161214_gel_Oly_gDNA.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20161214_gel_Oly_gDNA.jpg)
Overall, the gel looks OK. A fair amount of smearing, but a strong, high molecular weight band is present. The intensity of the smearing is likely due to the fact that the gel is overloaded for this particular well size. If I had used a broader comb and/or loaded less DNA, the band would be more defined and the smearing would be less prominent.
Will submit sample to the UW PacBio facility tomorrow!