We were previously approached by Cindy Lawley (Illumina Market Development) for possible participation in an Illumina product development project, in which they wanted to have some geoduck tissue and DNA on-hand in case Illumina green-lighted the use of geoduck for testing out the new sequencing platform on non-model organisms. Well, guess what, Illumina has give the green light for sequencing our geoduck! However, they need at least 4μg of gDNA, so I’m isolating more.
Isolated DNA from ctenidia tissue from the same Panopea generosa individual used for the BGI sequencing efforts. Tissue was collected by Brent & Steven on 20150811.
Used the E.Z.N.A. Mollusc Kit (Omega) to isolate DNA from five separate ~60mg pieces of ctenidia tissue according to the manufacturer’s protocol, with the following changes:
Samples were homogenized with plastic, disposable pestle in 350μL of ML1 Buffer
Incubated homogenate at 60C for 1hr
No optional steps were used
Performed three rounds of 24:1 chloroform:IAA treatment
Eluted each in 50μL of Elution Buffer and pooled into a single sample
Quantified the DNA using the Qubit dsDNA BR Kit (Invitrogen). Used 1μL of DNA sample.
Concentration = 162ng/μL (Quant data is here [Google Sheet]: 20170105_gDNA_geoduck_qubit_quant
Yield is great (total = ~32μg).
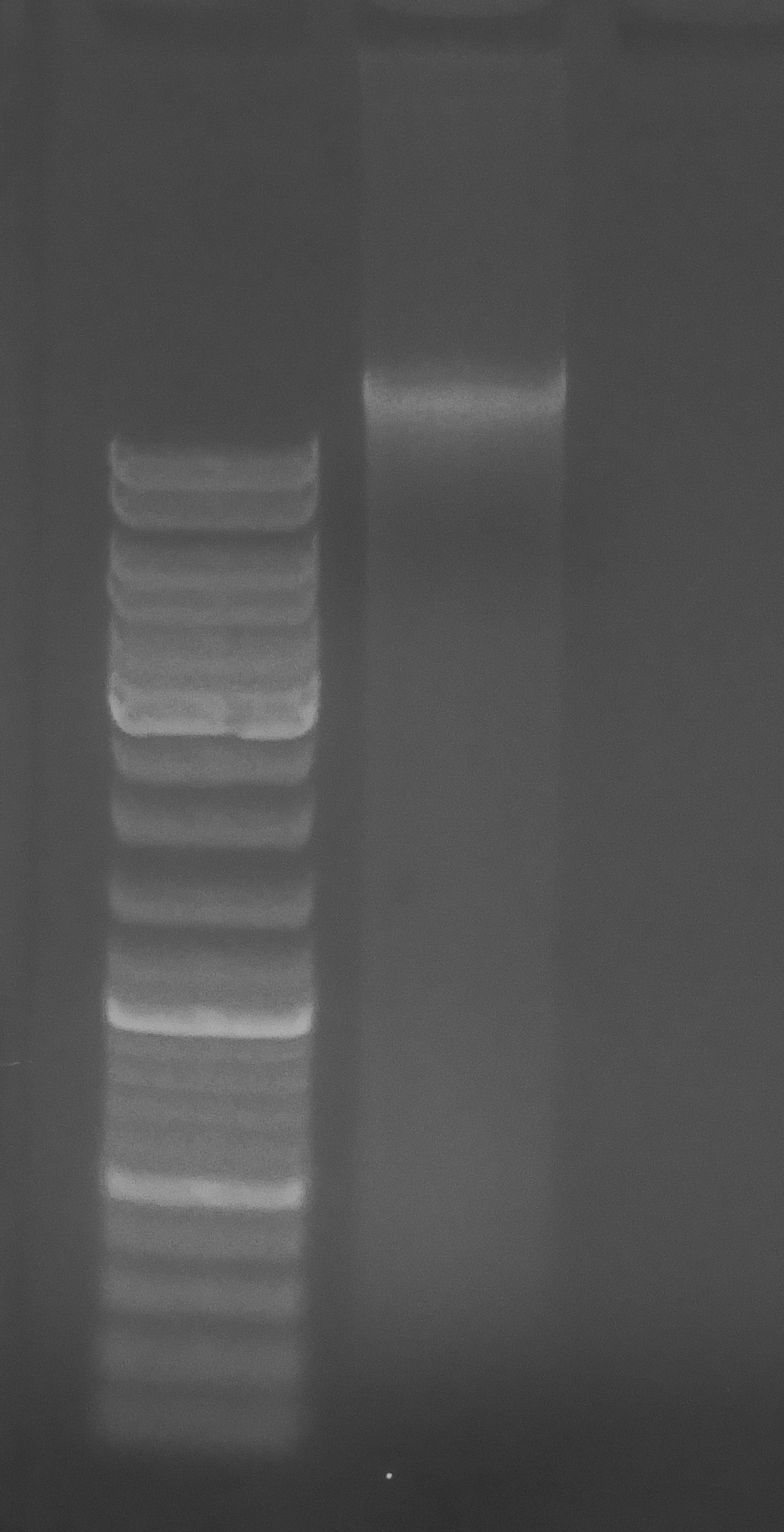
Evaluated gDNA quality (i.e. integrity) by running 162ng (1μL) of sample on 0.8% agarose, low-TAE gel stained with ethidium bromide.
Used 5μL of O’GeneRuler DNA Ladder Mix (ThermoFisher).
Results:
 (https://github.com/sr320/LabDocs/blob/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg?raw=true)
(https://github.com/sr320/LabDocs/blob/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg?raw=true)
 (http://eagle.fish.washington.edu/Arabidopsis/20170105_gel_geoduck_gDNA.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20170105_gel_geoduck_gDNA.jpg)
DNA looks good: bright high molecular weight band, minimal smearing, and minimal RNA carryover (seen as more intense “smear” at ~500bp).
Will send off 10μg (they only requested 4μg) so that they have extra to work with in case they come across any issues.