[I had previously submitted the two non-demultiplexed genotype-by-sequencing (GBS) files provided by BGI to the NCBI short read archive (SRA)(2017/02/08/data-management-sra-submission-of-ostrea-lurida-gbs-fastq-files.html).
He noticed that the SRA no longer wants “raw data dumps” (i.e. the type of submission I made before). So, that means I had to prepare the demultiplexed files provided by BGI to actually submit to the SRA.
Last week, I uploaded all 192 of the files via FTP. It took over 10hrs.
(FTP tips: - Use ftp -i to initiate FTP. - Then use open ftp.address.IP to connect. - Then can use mput with regular expressions to upload multiple files)
Today, I created a batch BioSample submission:
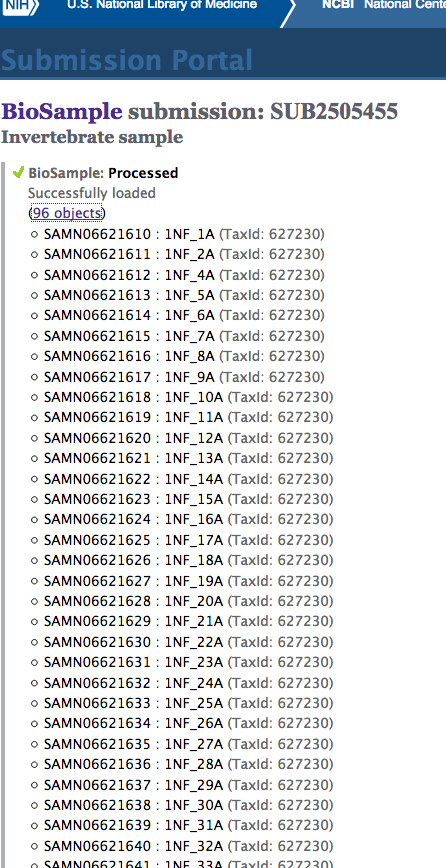 (http://eagle.fish.washington.edu/Arabidopsis/20170320_SRA_oly_gbs_demultiplex_SUB2505455.png)
(http://eagle.fish.washington.edu/Arabidopsis/20170320_SRA_oly_gbs_demultiplex_SUB2505455.png)
Initiated the submission process (Ummm, this looks like it’s going to take awhile…):
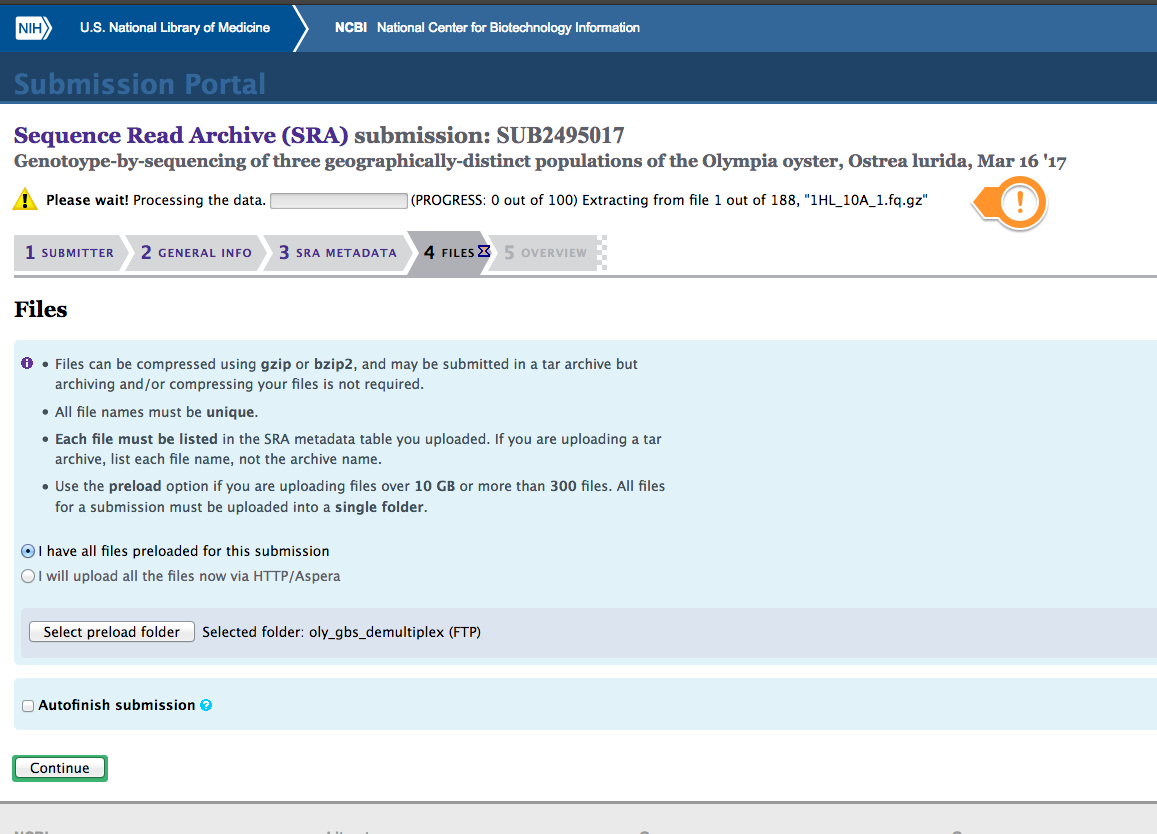 (http://eagle.fish.washington.edu/Arabidopsis/20170320_SRA_oly_gbs_demultiplex_SUB2495017.png)
(http://eagle.fish.washington.edu/Arabidopsis/20170320_SRA_oly_gbs_demultiplex_SUB2495017.png)
Aaaaaand, it failed:
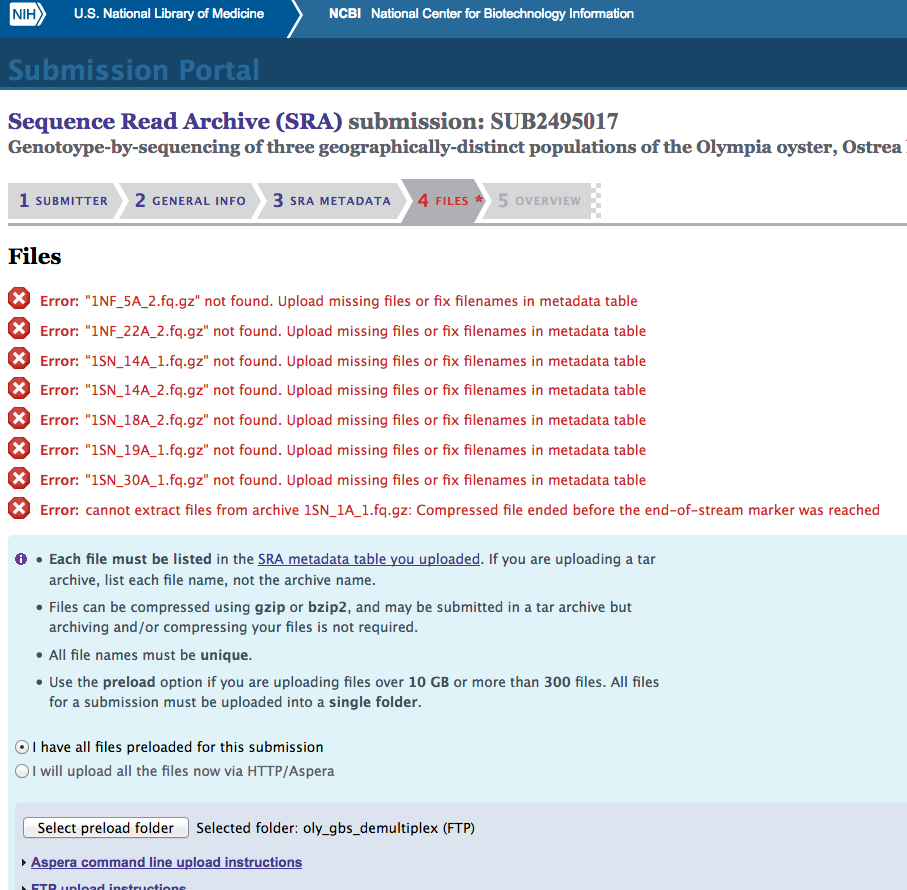 (http://eagle.fish.washington.edu/Arabidopsis/20170320_SRA_oly_gbs_demultiplex_SUB2495017_fail.png)
(http://eagle.fish.washington.edu/Arabidopsis/20170320_SRA_oly_gbs_demultiplex_SUB2495017_fail.png)
It seems like the FTP failed at some point, as there’s nothing about those seven files that would separate them from the remaining 185 files. Additional support for FTP failure is that the 1SN_1A_1.fq.gz error message makes it sound like only part of the file got transferred.
I’ll retrieve those files from our UW Google Drive (since their original home on Owl is still down) and get them trasnferred to the SRA.