I recently finished an assembly of our Olympia oyster PacBio data using Canu and thought it would be interesting to compare to Sean’s Canu assembly.
Granted, this isn’t a totally true comparison because I think Sean’s assembly is further “polished” using Pilon or something like that, but the Quast analysis is so quick (like < 60 seconds), that it can’t hurt.
See the Jupyter Notebook below for the full deets on running Quast.
Results:
Quast output folder: https://owl.fish.washington.edu/Athaliana/quast_results/results_2017_10_23_18_01_25/
Interactive report: https://owl.fish.washington.edu/Athaliana/quast_results/results_2017_10_23_18_01_25/report.html
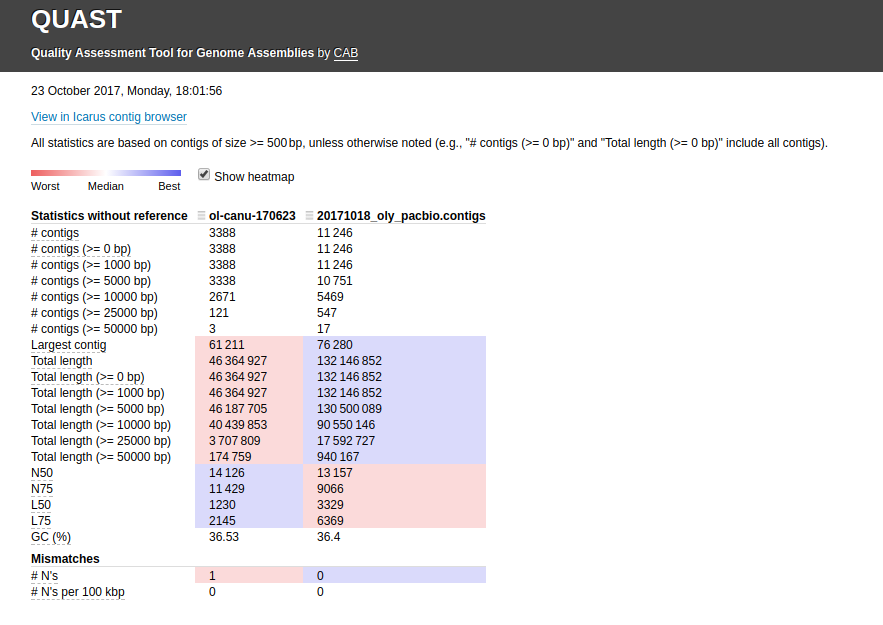 (http://owl.fish.washington.edu/Athaliana/20171023_quast_sbcanu_sjwcanu.png)
(http://owl.fish.washington.edu/Athaliana/20171023_quast_sbcanu_sjwcanu.png)
Jupyter Notebook (GitHub): 20171023_docker_oly_pacbio_canu_comparisons.ipynb