Earlier today, I ran kmergenie on our all of geoduck DNA sequencing data to see what it would spit out for an ideal kmer setting, which I would then use in another assembly attempt using SparseAssembler; just to see how the assembly might change.
The output from that kmergenie run suggested that the ideal kmer size exceeded the default maximum (k = 121), so I decided to run kmergenie a few more times, with some slight changes.
All jobs were run on our Mox HPC node.
Run 1
Diploid
Slurm script: 20180419_kmergenie_diploid_geoduck_slurm.sh
Run 2
Diploid
k 301
Slurm script: 20180419_kmergenie_diploid_k301_geoduck_slurm.sh
Results:
Output folders:
Slurm output files:
Kmer histogram (HTML) reports:
Diploid
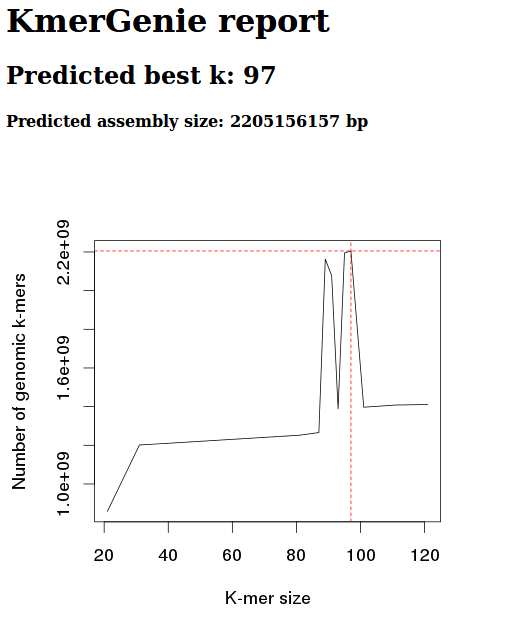
Diploid, k 301
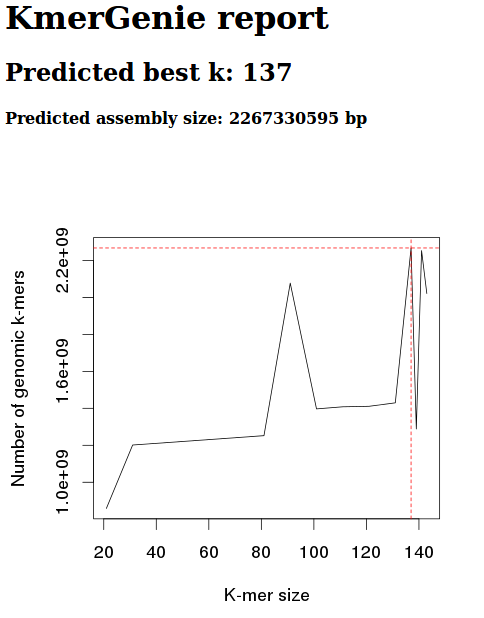
Okay, well, these graphs clearly show that the diploid setting is no good.
We should be getting a nice, smooth, concave curve.
Will try running again, without diploid setting and just increasing the max kmer size.