UPDATE 20191125
Since the results I obtained on my final attempt to get this to work failed, I decided to double-check the primer sequences.
Well, I ordered/used the wrong sequences! The two general Crassostrea spp. primers ordered were the 28s primers listed in that paper, instead of the cytochrome oxidase primers! I’ve ordered the correct universal CO primers, which are actually listed in this paper:
Will re-run this.
I’m leaving the original post below for posterity.
After yesterday’s PCR debacles, I re-ran the PCRs with the original cylcing parameters, on a long 1.5% agarose (1x low TAE) gel.
Primers and cycling parameters were taken from this publication:
| SR ID | Primer Name | Sequence |
|---|---|---|
| 1727 | COreverse | CAGGGGGCCGTTCGCGGTCAACGCT |
| 1726 | COCsi546r | AAGTAACCTTAATAGATCAGGGAACC |
| 1725 | COCgi269r | TCGAGGAAATTGCATGTCTGCTACAA |
| 1724 | COforward | GGGACTACCCCCTGAATTTAAGCAT |
This is a multiplex PCR, where the COforward/reverse primers should amplify any Crassostrea spp. DNA (i.e. a positive control - 697bp) and the other two primers will amplify either C.gigas (Cgi269r - 269bp) or C.sikamea (Csi546r - 546bp).
Master mix calcs:
| Component | Single Rxn Vol. (uL) | Num. Rxns | Total Volumes (uL) |
|---|---|---|---|
| DNA | 4 | NA | NA |
| 2x Apex Master Mix | 12.5 | 18 | 225 |
| COforward (100uM) | 0.15 | 18 | 2.7 |
| COreverse (100uM) | 0.15 | 18 | 2.7 |
| COCgi269r (100uM) | 0.1 | 18 | 1.8 |
| COCsi546r (100uM) | 0.1 | 18 | 1.8 |
| H2O | 8 | 18 | 144 |
| 25 | Add 21uL to each PCR tube |
Cycling params:
95oC for 10mins
30 cycles of:
- 95oC 1min
- 51oC 1min
- 72oC 1min
72oC 10mins
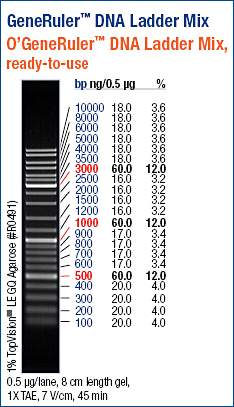
Used the GeneRuler DNA Ladder Mix (ThermoFisher) for all gels:
RESULTS
Gel:
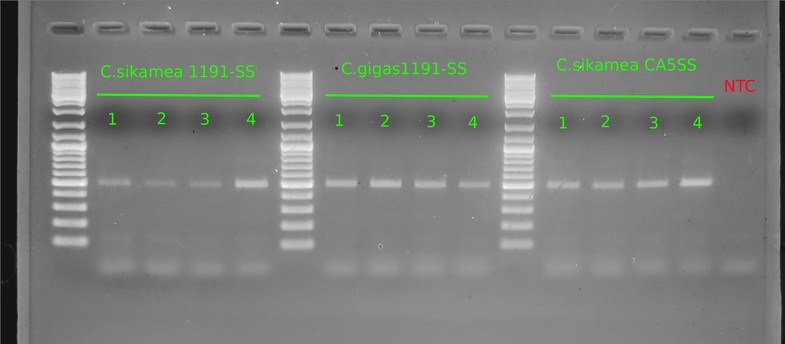
Well, despite the very clean appearance of this gel image (defined bands, no bands in NTC), the results are not helpful.
Band of ~700bp should be present in all samples (OCforward/reverse primers should amplify any Crassostrea spp DNA)- it isn’t present in any of them.
The single band generated in each lane is ~500 - 500bp. This band size is relatively close to the expected product size for Crassostrea sikamea detection (546bp).
These results could suggest that they actually all are C.sikamea. However, the C.gigas 1191-SS are supposed to be verified C.gigas; the samples in question were the C.sikamea 1191-SS.
Will discuss with Steven to see how much additional time he’d like to devote to this project to determine if I should re-run each of these samples with the species-specific primers only (i.e. no multiplex).