After noticing that the initial MEGAN6 taxonomic assignments for our combined C.bairdi NanoPore data from 20200917 revealed a high number of bases assigned to E.canceri and Aquifex sp., I decided to explore the taxonomic breakdown of just the individual samples to see which of the samples was contributing to these taxonomic assignments most.
20102558-2729-Q7 on 20200928: uninfected muscle
6129-403-26-Q7 on 20200928: Hematodinium-infected hemolymph
After completing the individual taxonomic assignments, I compared the two sets of assignments using MEGAN6 and generated this bar plot showing percentage of normalized base counts assigned to the following groups within each sample:
Aquifex sp.
Arthropoda
E.canceri
SAR (Supergroup within which Alveolata/Hematodinium sp. falls)
IMPORTANT!!!
- The taxonomic makeup shown in these comparisons is only a comparison of bases assigned amongst the four taxa selected above. It is not a comparison of the full taxonomic makeup of the two samples. I will discuss the data shown here in that context.
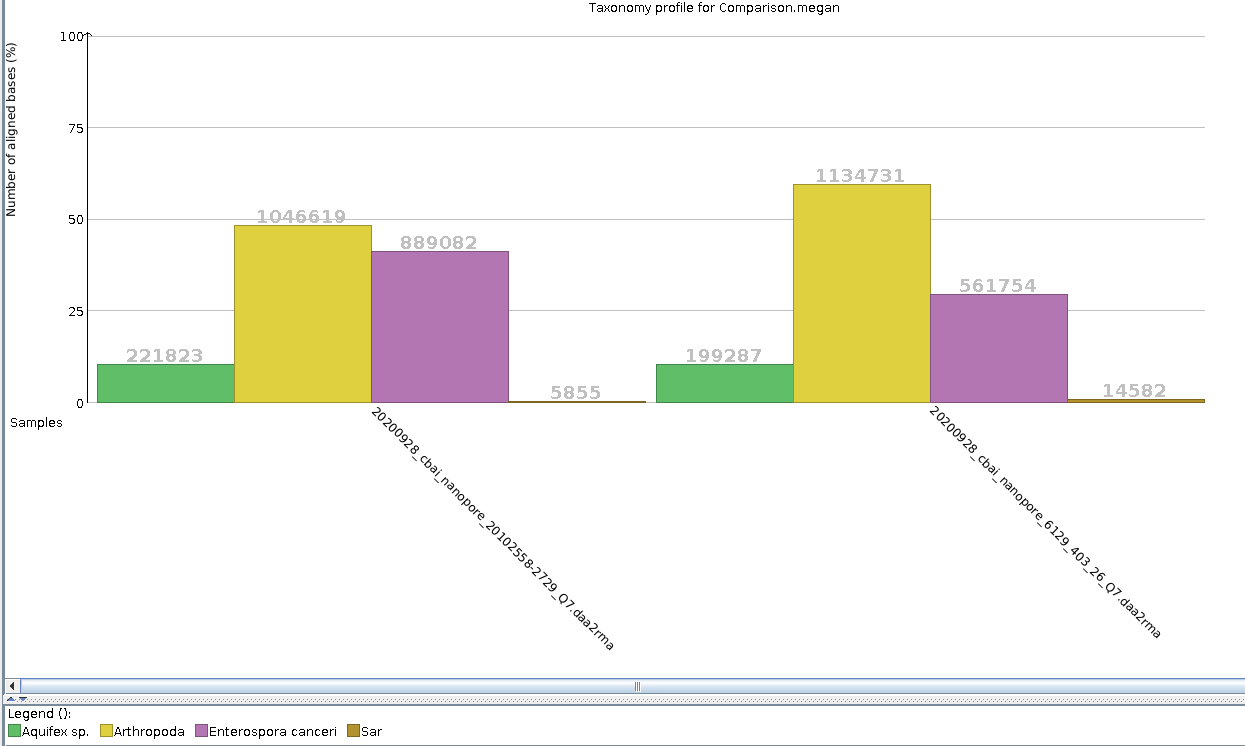
Comparison table:
| Taxa | 20102558-2729-Q7_base-counts | 20102558-2729-Q7_base-counts(%) | 6129-403-26-Q7_base-counts | 6129-403-26-Q7_base-counts(%) |
|---|---|---|---|---|
| Aquifex sp. | 221,823.00 | 10.25 | 199,287.06 | 10.43 |
| Arthropoda | 1,046,619.00 | 48.38 | 1,134,731.00 | 59.40 |
| Enterospora canceri | 889,082.00 | 41.10 | 561,754.19 | 29.41 |
| Sar | 5,855.00 | 0.27 | 14,582.56 | 0.76 |
| TOTAL | 2,163,379.00 | 1,910,354.81 |
Some observations:
Aquifex sp. account for nearly the same percentage of assignments in both samples.
Arthropoda makes up ~50% of assigned bases in the uninfected muscle sample (20102558-2729), but ~60% in the Hematodinium-infected hemolymph sample. (6129-403-26).
E.canceri makes up ~41% of assigned bases in the uninfected muscle sample (20102558-2729), but only ~30% in the Hematodinium-infected hemolymph sample.
SAR contributes a very small percentage to each of the two samples, but has ~2.8x the number of assigned bases. Additionally, as noted in the taxonomic assignment analysis of 20102558-2729-Q7 on 20200928, no bases are assigned to descendants of this Supergroup, whereas in the taxonomic analysis of 6129-403-26-Q7 on 20200928, there are bases assigned within the descendants of this Supergroup, down the level of Hematodinium sp. Genus.
Pretty interesting stuff!
I also briefly looked at the taxonomic assignments from all of our hemolymph RNAseq samples to see if if Aquifex sp. and/or E.canceri appear:
Interestingly, a high number of reads are assigned to E.canceri in all samples, but no reads are assigned to Aquifex sp.. Another observation is that a fair number of reads get assigned to Vibrio parahemolyticus, but very few number of NanoPore DNA bases get assigned to V.parahemolyticus.
Next up I think I might try to identify which contigs/scaffolds from the cbai_genome_v1.0 Flye assembly correspond to these taxa. The approach would be to create a BLAST database (DB) from the cbai_genome_v1.0.fasta (19MB). Then extract the NanoPore reads assigned to each of the taxa above, then BLAST them against the cbai_genome_v1.0 BLAST DB.