Steven assigned me to do some MBD-BSseq library prep (GitHub Issue) for some Dungeness crab (Metacarcinus magister) DNA samples provided by Mackenzie Gavery. The DNA was isolated from juvenile (J6/J7 developmental stages) gill tissue. One of the first steps in MBD-BSseq is to fragment DNA to a desired size (~350 - 500bp in our case). However, we haven’t worked with Metacarcinus magister DNA previously, so I need to empirically determine sonicator (Bioruptor 300; Diagenode) settings for these samples.
I used 1ug of DNA in a volume of 50uL, using 0.65mL prelubricated snap cap tubes (Costar; Cat# 3206).
Initially, I did a 35 cycle (30s ON, 30s OFF; low intensity setting) run and determined it was insufficient, so ran increments of 5 cycles and pulled out 1.5uL after each one to run on the Bioanalyzer.
Post-sonication/shearing, samples were run on High Sensitivity DNA Assay chips in the Bioanalyzer 2100 (Agilent).
RESULTS
Output folder/files:
-
Bioanalyzer files (XAD; require 2100 Expert software to open)
Quick summary: Potentially need to cycle for 70 cycles (!!!!) to achieve desired average fragment length.
Will test an uninterrupted run tomorrow to confirm that these settings are accurate.
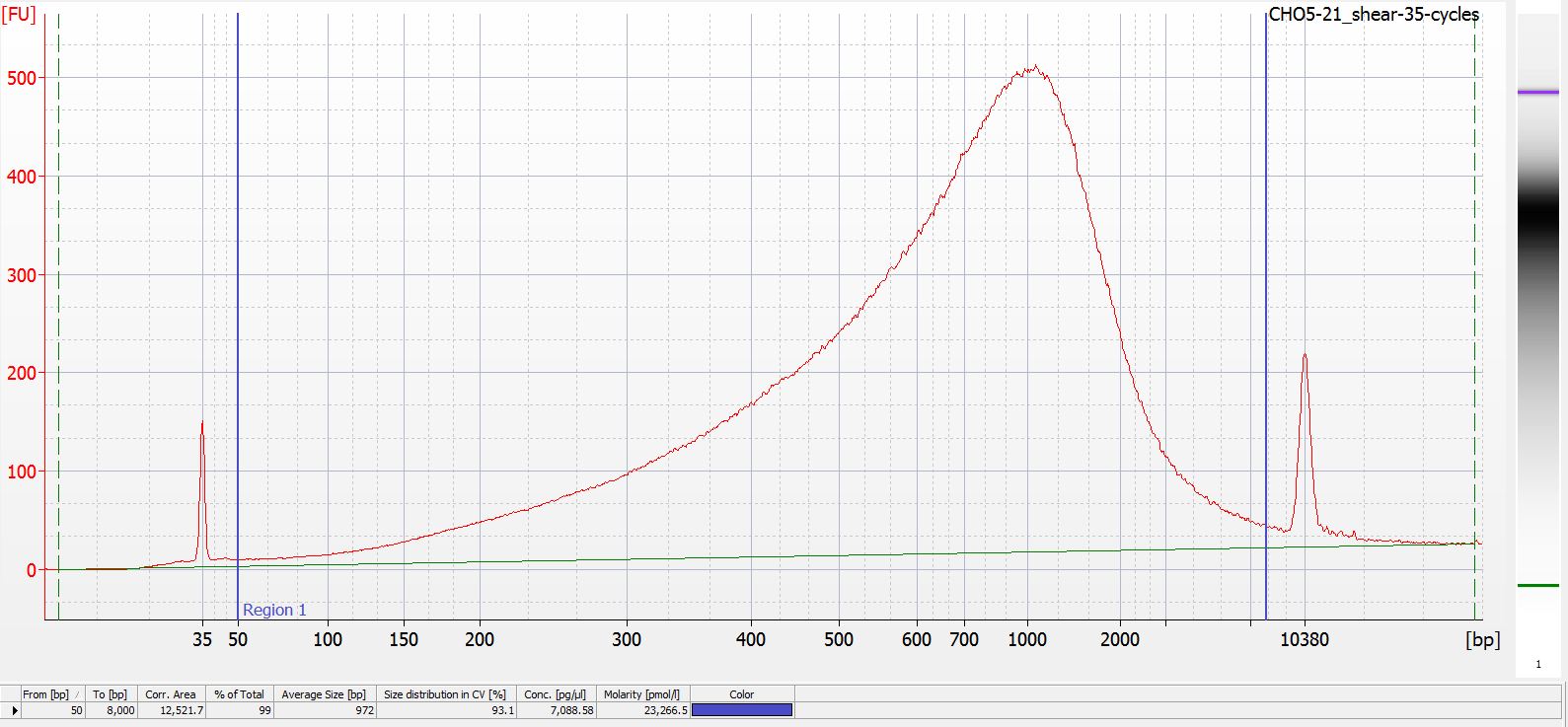
Below are all of the electropherograms from each of the total cycles the CHO5-21 gDNA was subjected to: 35 - 80 cycles, increments of 5 cycles. Directly underneath each electropherogram (it’s small - you’ll need to open the image in a new tab to see better) is data showing the mean fragment size within the regions of 50 - 8000bp for each sample.
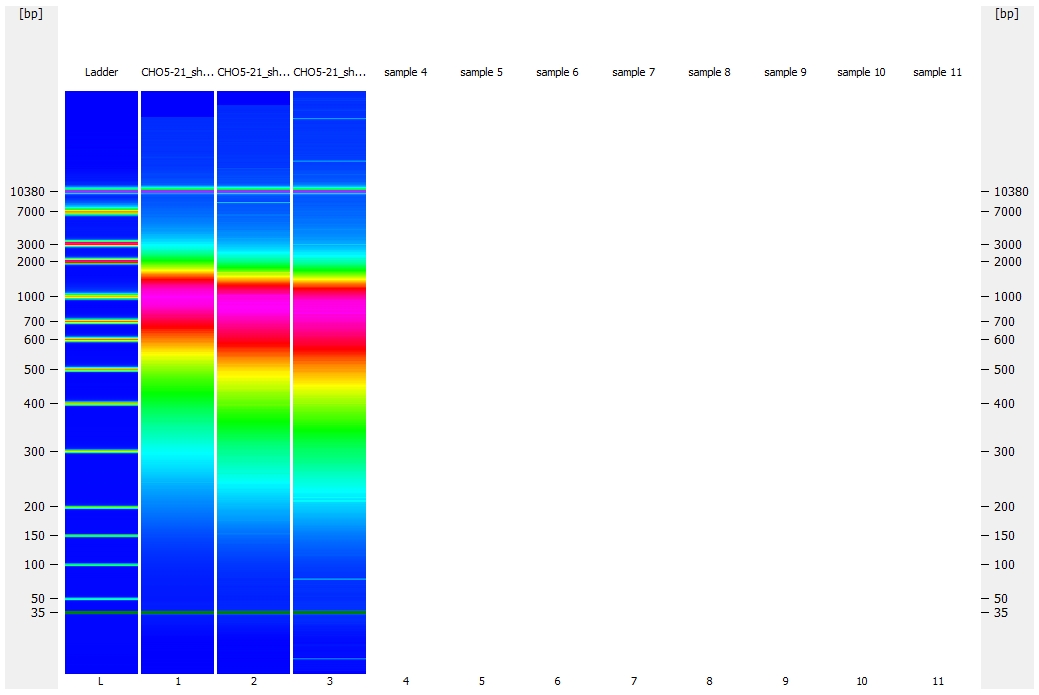
There are also two “pseudo-colored” (this is the software verbiage for heatmap-like coloration) gel representation comparisons of samples. It’s an easy way to see the progression of fragmentation as the cycle number increases.
Also, I ended up running three chips (gah!) to accommodate all of this (was not expecting to need to fragment for this number of cycles) and that is why there is not a singular gel representation image allowing for the comparison of all the samples together.
Electropherogram: 35 cycles
Electropherogram: 40 cycles
Electropherogram: 45 cycles
Electropherogram: 50 cycles
Gel comparison: 40, 45, and 50 cycles