After generating a new Ostrea lurida genome assembly (v090) on 20210520, I decided to compare with our previous genome assemblies. Here, I compared v090 with all of our previous scaffolded assemblies using Quast. Here is a table (GitHub) which describes all of our existing assemblies (i.e. assembly name, assembly process, etc):
NOTE: Assembly pbjelly_sjw_01 is the source of our current “canonical” assembly: Olurida_v081.fa
This was run locally on my computer with the following command:
python \
/home/sam/programs/quast-5.0.2/quast.py \
--threads=20 \
--min-contig=100 \
--labels=Olurida_v090,canu_sb_01,canu_sjw_01,platanus_sb_01,platanus_sb_02,racon_sjw_01 \
~/data/O_lurida/genomes/Olur_v090.SPolished.asm.wengan.fasta \
/mnt/owl/scaphapoda/Sean/Oly_Canu_Output/oly_pacbio_.contigs.fasta \
/mnt/owl/Athaliana/20171018_oly_pacbio_canu/20171018_oly_pacbio.contigs.fasta \
/mnt/owl/scaphapoda/Sean/Oly_Illumina_Platanus_Assembly/Oly_Out__contig.fa \
/mnt/owl/scaphapoda/Sean/Oly_Platanus_Assembly_Kmer-22/Oly_Out__contig.fa \
/mnt/owl/Athaliana/201709_oly_pacbio_assembly_minimap_asm_racon/20170918_oly_pacbio_racon1_consensus.fastaRESULTS
Output folder:
20210521_olur_quast_non-scaffold_assembly-comparisons/
Quast Report (HTML; open in browser; interactive)
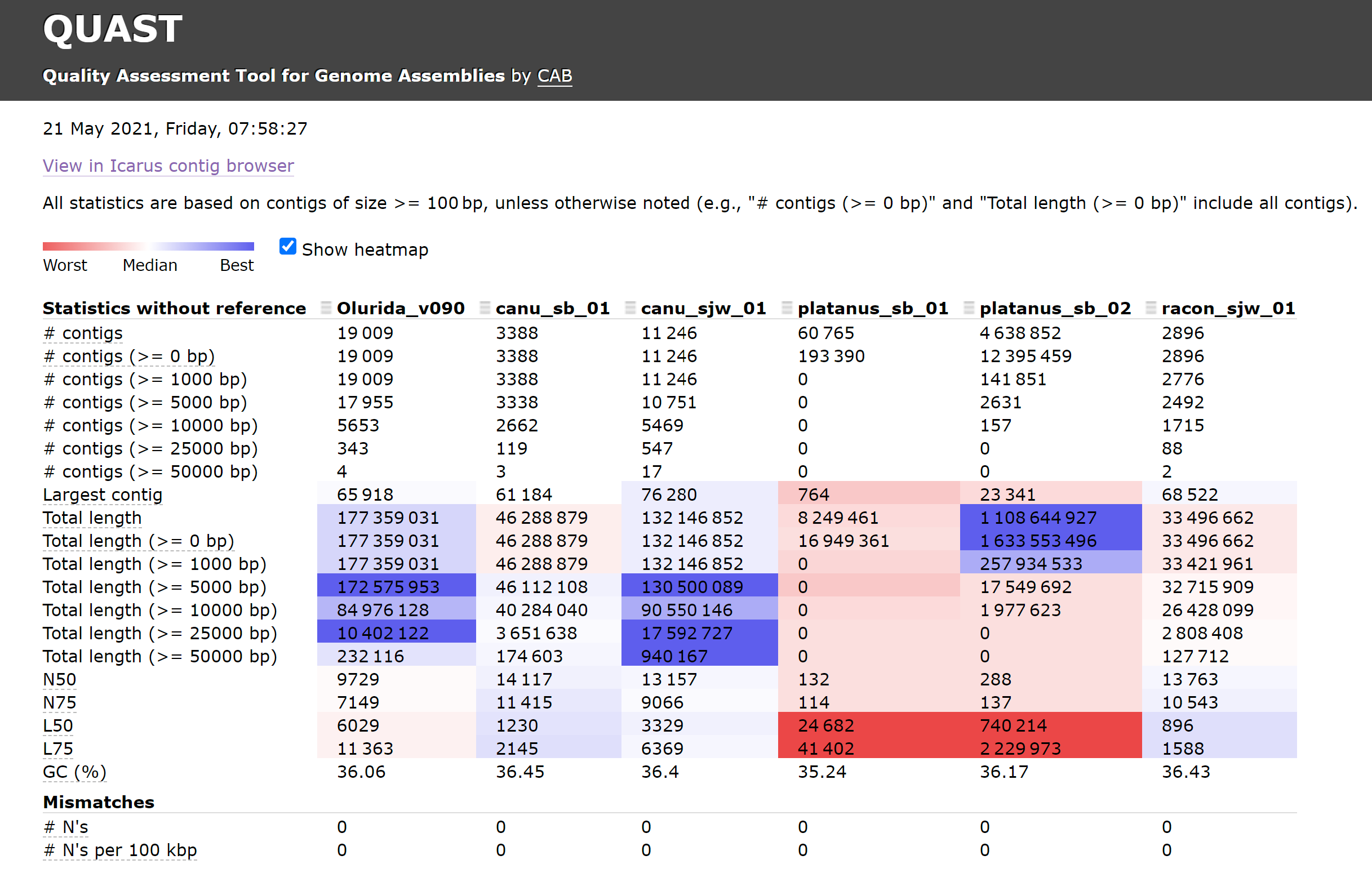
The results here are interesting. The Olurida_v090 assembly which I generated on 20210520 Looks pretty good. However, despite having ~44M fewer bases, the canu_sjw_01 assembly (from 20171018) could be considered “better”, as it has ~4x the number of contigs >50,000bp than the Olurida_v090. The canu_sjw_01 assembly also has the larger of the two assemblies’ largest contigs. Also, surprisingly, the canu_sjw_01 assembly is only our PacBio sequencing data; it does not include any of the Illumina short reads!
Maybe I’ll just go ahead and run both of these through GenSAS…