Alrighty, this notebook entry is going to have a lot to unpack, as the process to get these pipelines running and then deal with the actual data we wanted to run them with was quite involved. However, the TL;DR of this all is this:
Both EpiDiverse pipelines (
wgbsandsnp) are running properly on our computer, Raven.The Ostrea lurida (Olympia oyster) data which Steven wanted to identify SNPs in is possibly screwy?
Anyway, now to the meat of everything! If not interested in all the ins/outs, skip down to the Results section to see the various comparisons which were run. This analysis was spurred by this GitHub Issue. Steven wanted to run some Ostrea lurida (Olympia oyster) MBD BSseq data (from December 2015). These data are single-end, 50bp FastQs from ZymoResearch, with files named zr1394*. They were trimmed with TrimGalore on 20180503. Steven ran the trimmed data through Bismark and created deduplicated, sorted BAMs on 2020205. These BAMs were created using our Olurida_v081.fa.
Attempted to run the deduplicated, sorted BAMs through the EpiDiverse/snp Nextflow pipeline, but repeatedly encountered a memory error. Memory limitations were also an issue Steven had also encountered when trying to do the same anlalysis using BS-Snper. So, Steven created a reduced genome and ran the data through Bismark using that genome.
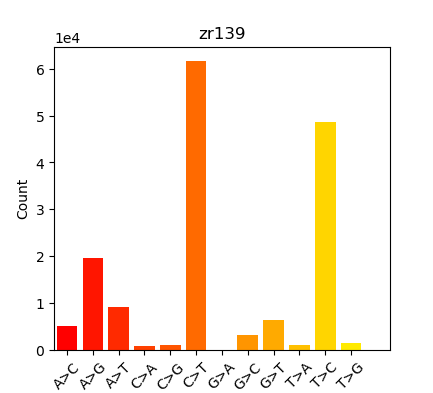
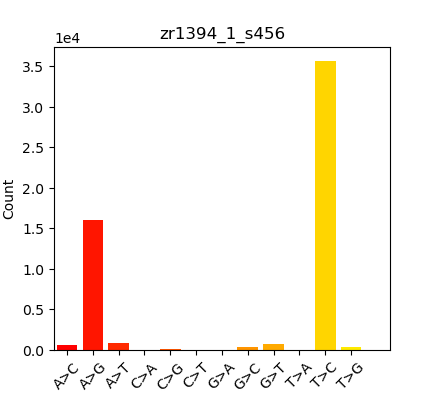
Was finally able to run those BAMs through the EpiDiverse/snp Nextflow pipeline, but the “Substitutions” plot looked like this:
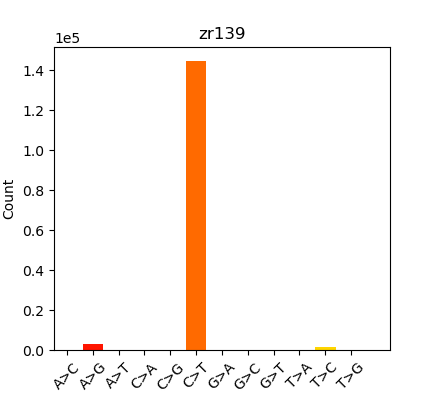
These results didn’t look like what we’d expected (expected a more equal distribution on SNPs…); which triggered a rabbit hole of exploration and testing. Performed the following runs to see if I could identify the source of this skewed SNP distribution:
Original Oly trimmed FastQs through
EpiDiverse/wgbs, followed byEpiDiverse/snp.Raw FastQs trimmed with default settings in the
EpiDiverse/wgbspipeline followed by theEpiDiverse/snppipelineRaw FastQs trimmed with additional hard clipping of 10bp from 5’ end to match how original trimming on 20180503 had been performed (trimming regimen is what’s recommended by
TrimGalorefor this type of data) using theEpiDiverse/wgbspipeline, followed byEpiDiverse/snppipeline
RESULTS
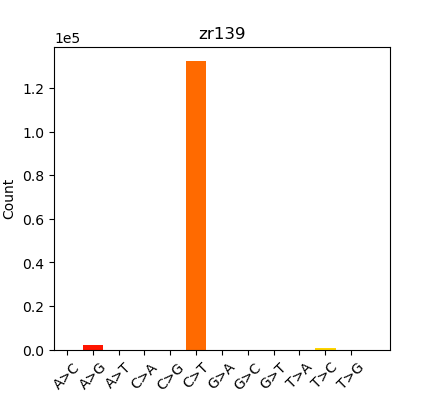
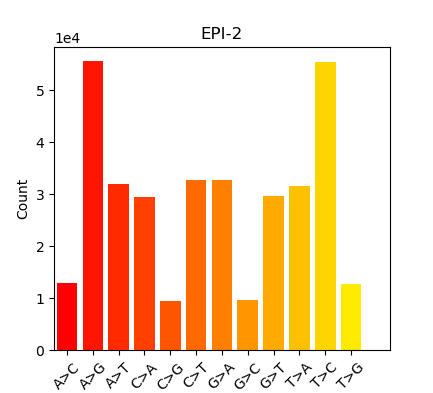
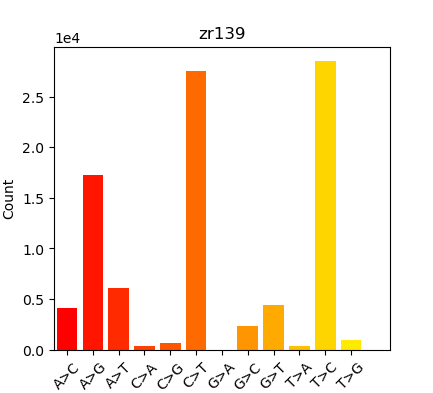
Summary in table form might be the easiest way to present this:
| Oly Bismark | Geoduck Bismark | Oly EpiDiverse Adaptor Trim | Oly EpiDiverse Adaptor and 10bp 5’ Trim | Oly EpiDiverse Adaptor and 10bp 5’/3’ trim |
|---|---|---|---|---|
 |
 |
 |
 |
 |