20240229
Lab Meeting
- Discussed sections from “Braiding Sweetgrass”
CEABIGR (GitHub repo)
Finalized females/males controls/exposed expression table.
Weekly meeting.
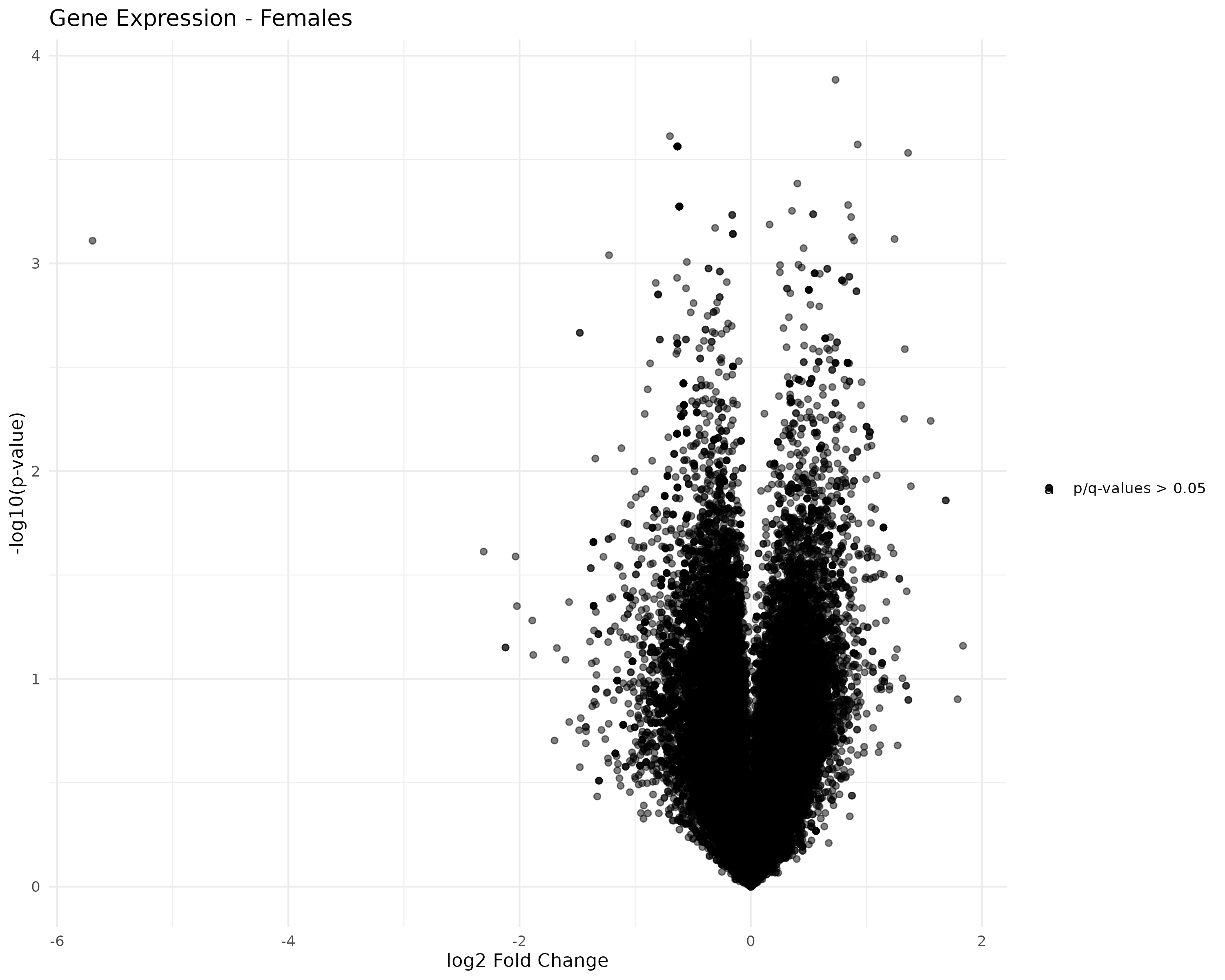
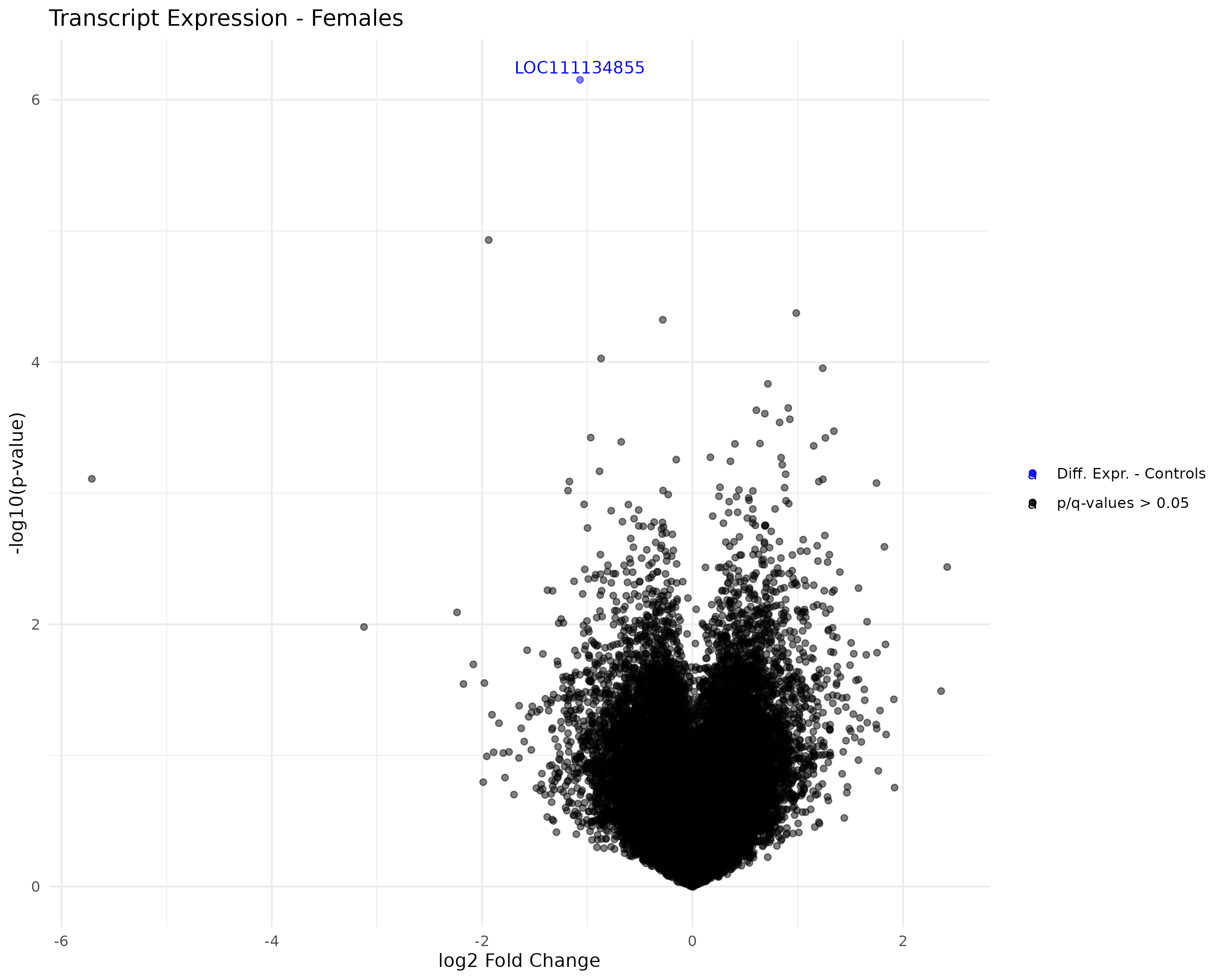
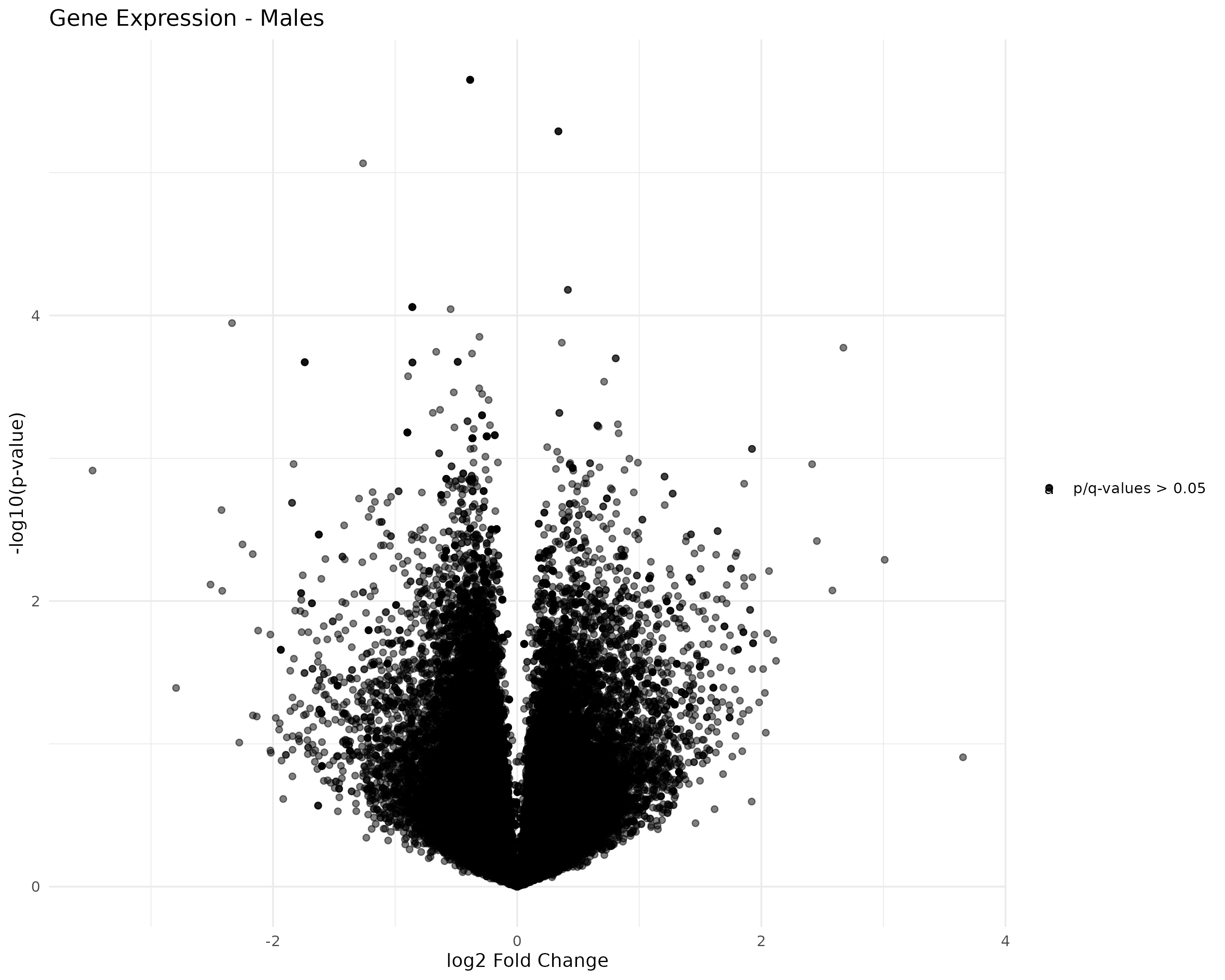
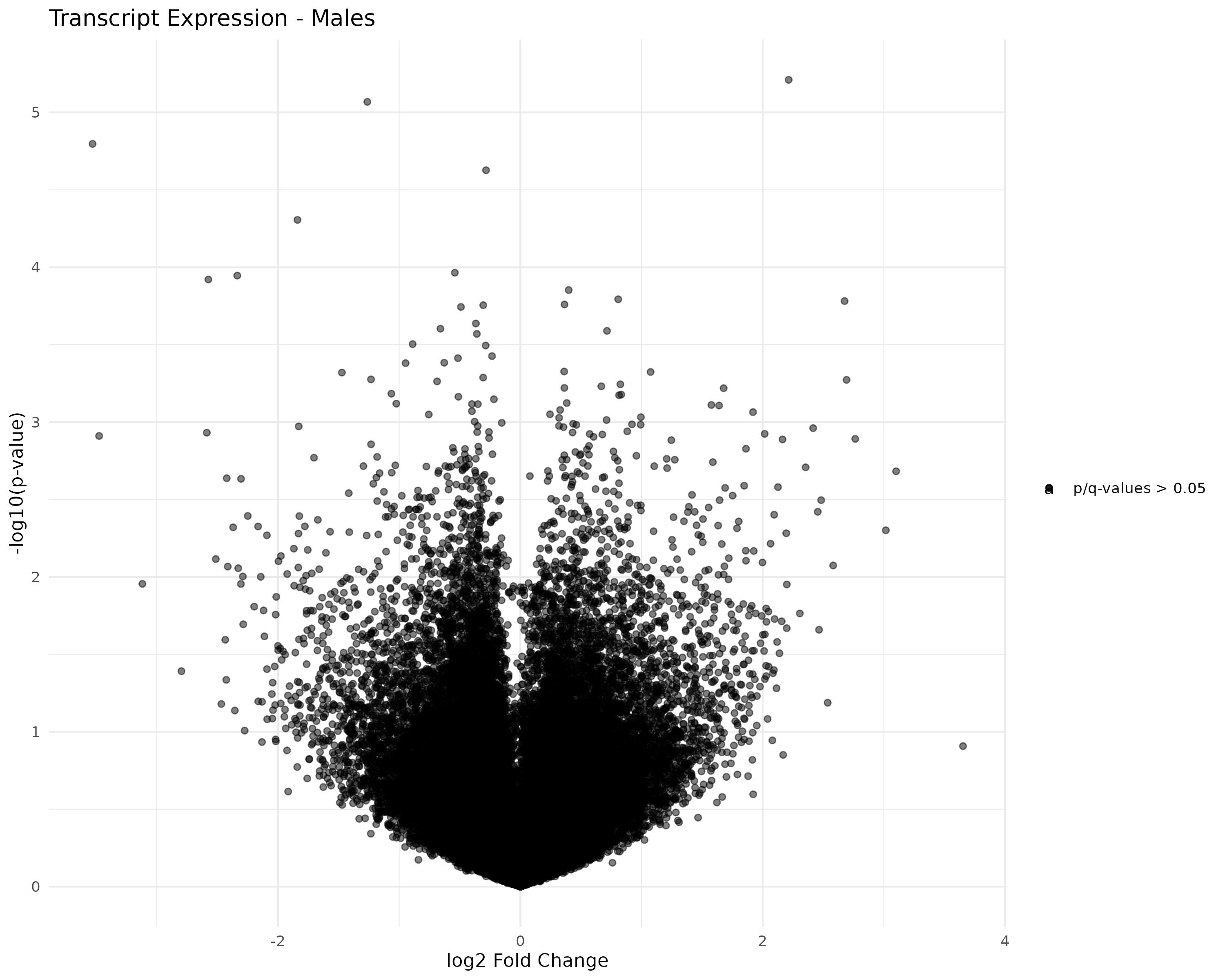
Created faceted volcano plots of female/male control/exposed transcript and gene expression:

20240228
Miscellaneous
- Read designated sections of “Braiding Sweetgrass” for lab meeting.
CEABIGR (GitHub repo)
- Created supplemental table with female/male controls/exposed expression info.
20240227
Miscellaneous
Setup account on Owl for Ariana. GitHub Issue
Set up access to nightingales Google Sheet for Ariana. GitHub Issue
CEABIGR (GitHub repo)
- Generated table with mean FPKM values for genes in female/male control/exposed.
20240226
In lab
Selected and prepared primers for Eric’s qPCRs.
Helped Eric conduct qPCRs.
Helped Katherine gather sampling supplies for hatchery.
Ran all over the place in Bagley Hall to get LN2! Chemistry Stores was out and so was secondary location in Rm. 305A. Eventually managed to get LN2 in 405A…
CEABIGR (GitHub repo)
- Generated canonical predominant isoform table.
20240223
Coral E5 (GitHub repo)
All hands meeting.
Read original proposal.
Read paper about plant epigenetic stress responses.
Science Hour
CEABIGR (GitHub repo)
- Created canonical max transcripts per gene table for females controls/exposed and males controls/exposed.
20240222
Lab Meeting
- Discussed Pub-a-thon 2024
CEABIGR (GitHub repo)
Reviewed recent GitHub Issues
Worked on creating canonical gene/transcript counts table.
Miscellaneous
Updated
Nightingales(Google Sheet) spreadsheet with Cod RNA-seq info.Raven was bogged down by some weird rogue
gitcommand, so had to reboot.
20240221
Miscellaneous
Set Ariana up with Gannet access (GitHub Issue)
Received Pacific cod RNA-seq data
CEABIGR (GitHub repo)
- Generated volcano plots
20240220
CEABIGR (GitHub repo)
- Started working on generating volcano plots.
20240216
Coral E5 (GitHub repo)
- Finished running
ShortStackvariations with newly trimmed P.meandrina sRNA-seq.
20240215
Coral E5 (GitHub repo)
- Ran
ShortStackvariations with newly trimmed P.meandrina sRNA-seq.
20240214
Coral E5 (GitHub repo)
- Ran
ShortStackvariations with newly trimmed Acropora pulchra sRNA-seq.
20240213
Coral E5 (GitHub repo)
- Cranked out new trimming on P.meandrina sRNA-seq samples.
20240212
In lab
- Reviewed reverse transcription and qPCR with Eric.
Coral E5 (GitHub repo)
- Started running new trimming on A.pulchra sRNA-seq samples (preivously, had just tried with P.evermanni).
20240209
Science Hour
- Steven and I looked at and discussed the exon expression patterns of individual genes and compared those patterns to the WGCNA outputs which Ariana generated at yesterday’s meeting. Definitely a few individuals whose exon usage seems to deviate a great deal from that WGCNA module assignment…
Coral E5 (GitHub repo)
- Continued to work on implementing new trimming/merging parameters, but ShortStack keeps encountering an error (GitHub Issue). I’m hoping the developer can lend some insight. This is weird, because these samples, albeit trimmed with a different program and used as paired-end reads (only trying R1 or merged reads at this point), were successfully analyzed previously…
20240208
Lab meeting
CEABIGR (GitHub repo)
- Special, extended session with Arianna in attendance. She ran some WGCNA (PDF) analysis to try to see how exon expression patterns varied between control/exposed samples. Very cool!
20240207
Coral E5 (GitHub repo)
Finished messing around with trimming/merging stuff. Here’s the notebook entry.
Began implementing new trimming/merging.
20240206
Coral E5 (GitHub repo)
- Continued to explore trimming and merging strategies for sRNA-seq data, using
fastpalone, BBMerge alone, and/or combining the two approaches.
CEABIGR (GitHub repo)
- Spent a few hours on a video chat with Steven and Yaamini going through exon fold-change code/analysis.
20240205
Coral E5 (GitHub repo)
- Continued to explore trimming and merging strategies for sRNA-seq data, using
fastpalone, BBMerge alone, and/or combining the two approaches.
20240202
Coral E5 (GitHub repo)
Science Hour
- Steven and I played around with exon fold-change data.
20240201
In Lab
Lab meeting.
Helped Eric get started with RNA quantification using Qubit.
Coral E5 (GitHub repo)
Tested out more R1/R2 merging/trimming using
fastp.- Merging is similar to BBMerge results - i.e. a (relatively) small number of reads are actually merging, despite the fact that these should be short insert sizes and R1/R2 reads should overlap…
Installed XICRA (Sanchez Herrero et al. 2021) on Raven to identify isomiRs in sRNA-seq data. Possibly to get around the merging “issues” experienced with BBMerge and
fastp.Installation was a bit bumpy, due to some weird Conda/Mamba “flexible solve” problem, but got that straightened out with
conda config --set channel_priority flexible.Installation test(s) had issues. I created the following GitHub Issues on the developer’s site in hopes of getting help:
CEABIGR (GitHub repo)
- Need to generate R2 values and slope of exon expression for all genes, Exons 1 - 6.