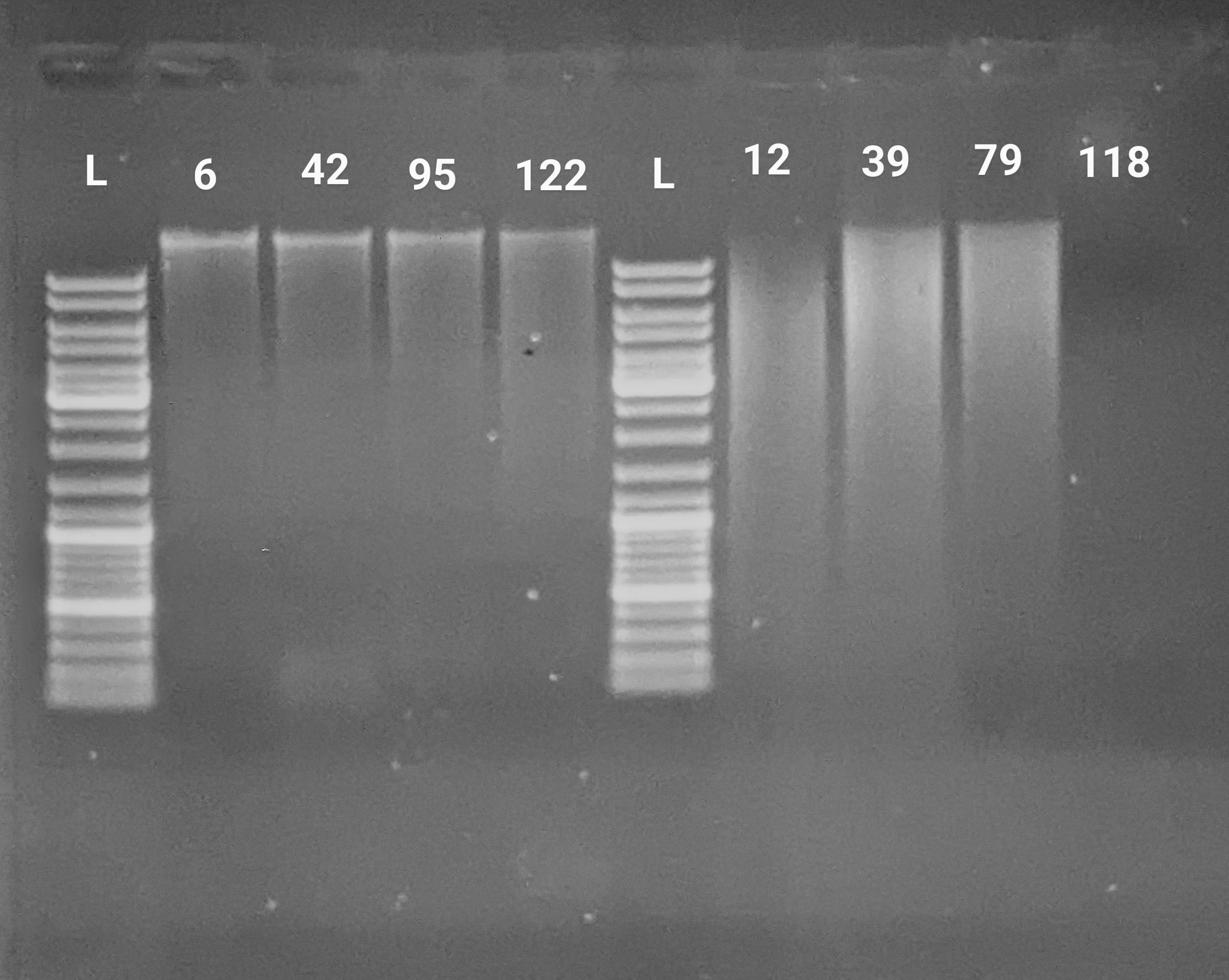
INTRO
Previously isolated and quantified gDNA on 20240531 (Notebook) and needed to evaluate gDNA integrity. We recently sent a bunch of liver samples off to Psomagagen for whole genome bisulfite sequencing (WGBS). As documented in this GitHub Issue, Psomagen discovered that the DNA quality from the samples was poor (i.e. heavily degraded). Steven asked to test DNA isolations on our end with four untouched samples, and four of the samples sent to Psomagen:
Untouched liver samples (never sent for RNA or DNA extraction), one from each treatment: 6, 42, 95, 122
Liver samples that were sent for DNA extraction for which we still have tissue, one from each treatment: 12, 39, 79, 118
Sample 118 did not have any tissue in the tube.
METHODS
Prepared 100mL of 0.8% agarose in 1x Low TAE, with ethidiumn bromide and 12-well, 1.5mm comb.
DNA was diluted to 20ng/uL, in a volume of 5uL. One microliter of 6X Orange DNA Loading Dye (ThermoFisher) was mixed with each diluted DNA sample and loaded in the gel.
Five microliters of Thermo Scientific™ O’GeneRuler DNA Ladder Mix, Ready-to-Use 100-10,000 bp was loaded in the gel.
The gel was run at 100V for ~1.5hrs.
RESULTS
The untouched samples look as one would expect - defined, high-molecular weight band with minimal smearing. This is indicative of high quality, intact gDNA. However, the previously touched samples show signs of heavy degradation, as evidenced by a lack of defined, high-molecular weight band, as well as extensive smearing.