- 1 CREATE OUTPUT DIRECTORY
- 2 edgeR GROUPING VECTOR
- 3 DEGs
- 3.1 Load count matrix
- 3.1.1 Reformat header
- 3.1.2 Convert to matrix
- 3.1.3 Load matrix into edgeR
- 3.1.4 Filter low counts
- 3.1.5 Normalization
- 3.1.6 Model fitting and estimating dispersions
- 3.1.7 Exact test (Gene expression)
- 3.1.8 Differential gene expression summary
- 3.1.9 Peek at DEGs
- 3.1.10 Convert gene ID rownames to column
- 3.1.11 Write DEGs to file
- 3.1.12 Volcano plot
- 3.1 Load count matrix
- 4 CITATIONS
Differential gene expression of 9oC vs 16oC liver RNA-seq using edgeR (Chen et al. 2024; McCarthy, Chen, and Smyth 2012; Chen, Lun, and Smyth 2016; Robinson, McCarthy, and Smyth 2009)
Note
Copied from 13.0.0-RNAseq-edgeR.md, commit f5fa415.
1 CREATE OUTPUT DIRECTORY
# Make output directory, if it doesn't exist
mkdir --parents ../output/13.0.0-RNAseq-edgeR2 edgeR GROUPING VECTOR
2.1 Check sample info
Only looking at 9oC and 16oC
echo "Header for ../data/DESeq2_Sample_Information.csv:"
echo ""
head -n 1 ../data/DESeq2_Sample_Information.csv
echo ""
echo "---------------------------------------------"
echo ""
awk -F"," 'NR==1 {header=$2 " " $4; print header; next} $4 == 16 || $4 == 9 {print $2, $4}' ../data/DESeq2_Sample_Information.csv \
| sort -k1,1 -n \
| column -tHeader for ../data/DESeq2_Sample_Information.csv:
sample_name,sample_number,tank,temp_treatment,tissue_type
---------------------------------------------
sample_number temp_treatment
1 16
2 16
3 16
4 16
5 16
10 16
11 16
12 16
13 16
18 16
19 16
19 16
19 16
20 16
20 16
20 16
21 16
28 16
29 16
30 16
31 16
36 16
78 9
79 9
80 9
83 9
88 9
90 9
91 9
92 9
94 9
97 9
98 9
99 9
100 9
107 9
108 9
109 9
110 9
116 92.2 Create edgeR grouping vector
Since all sample IDs <= to 36 are part of the 16oC treatment, we can use this to create vector which matches sample ordering in ../output/10.1-hisat-deseq2/gene_count_matrix.csv
# Read the first line of the CSV file
header <- readLines("../output/10.1-hisat-deseq2/gene_count_matrix.csv", n = 1)
# Split the header to extract the values
values <- as.numeric(unlist(strsplit(header, ","))[-1]) # Remove the first element which is "gene_id"
# Apply the conditional logic to generate the temperatures vector
temperatures <- ifelse(values <= 36, "16C", "9C")
# Print the resulting vector
print(temperatures) [1] "16C" "16C" "9C" "9C" "9C" "9C" "16C" "9C" "9C" "16C" "16C" "16C"
[13] "16C" "16C" "16C" "16C" "16C" "16C" "16C" "16C" "16C" "16C" "16C" "16C"
[25] "9C" "9C" "9C" "9C" "9C" "9C" "9C" "9C" "9C" "9C" "9C" "9C" 3 DEGs
3.1 Load count matrix
3.1.1 Reformat header
# Read the entire CSV file
csv_file <- readLines("../output/10.1-hisat-deseq2/gene_count_matrix.csv")
# Extract the header line
header <- csv_file[1]
# Split the header to extract the values
header_values <- unlist(strsplit(header, ","))
# Prepend 'sample_' to each value (excluding 'gene_id')
header_values[-1] <- paste0("sample_", header_values[-1])
# Combine the modified header back into a single string
modified_header <- paste(header_values, collapse = ",")
# Replace the old header with the new modified header in the CSV content
csv_file[1] <- modified_header
# Convert the modified CSV content to a data frame
csv_content <- read.csv(textConnection(csv_file), row.names = 1)
str(csv_content)'data.frame': 30575 obs. of 36 variables:
$ sample_1 : int 360 694 0 10 0 0 0 402 3696 335 ...
$ sample_10 : int 464 325 10 22 9 40 3 550 2492 171 ...
$ sample_100: int 391 276 13 15 0 40 0 293 2615 149 ...
$ sample_107: int 346 77 11 19 0 29 0 182 2184 99 ...
$ sample_108: int 691 409 49 74 18 8 41 1082 2702 241 ...
$ sample_109: int 408 196 42 28 0 0 7 213 2579 149 ...
$ sample_11 : int 436 281 14 63 0 38 4 234 3631 286 ...
$ sample_110: int 509 284 36 35 0 0 0 149 3158 139 ...
$ sample_116: int 366 327 0 30 0 16 3 444 2121 100 ...
$ sample_12 : int 373 310 14 31 27 15 0 360 2673 168 ...
$ sample_13 : int 432 393 9 25 0 0 0 531 2893 267 ...
$ sample_18 : int 385 386 0 42 0 0 0 242 2210 205 ...
$ sample_19 : int 330 106 0 17 0 0 27 269 2324 139 ...
$ sample_2 : int 288 290 14 14 0 0 0 194 2384 201 ...
$ sample_20 : int 307 252 3 21 0 22 0 359 2561 123 ...
$ sample_21 : int 346 326 21 60 0 22 0 383 2654 215 ...
$ sample_28 : int 293 345 26 61 0 69 0 395 2887 157 ...
$ sample_29 : int 347 254 9 14 0 22 4 392 2250 101 ...
$ sample_3 : int 452 363 39 29 4 0 0 408 3143 222 ...
$ sample_30 : int 452 353 20 37 35 15 10 489 2830 183 ...
$ sample_31 : int 984 922 10 68 0 0 0 310 1297 270 ...
$ sample_36 : int 230 160 0 37 0 34 23 352 1981 93 ...
$ sample_4 : int 469 588 0 0 0 55 0 168 2971 290 ...
$ sample_5 : int 400 285 10 30 4 51 0 403 2651 225 ...
$ sample_78 : int 472 178 48 10 0 0 0 462 2669 195 ...
$ sample_79 : int 311 119 41 13 0 30 0 287 2671 182 ...
$ sample_80 : int 368 197 60 56 0 19 4 310 1790 123 ...
$ sample_83 : int 312 134 39 32 0 31 4 113 2173 162 ...
$ sample_88 : int 551 128 49 45 8 0 0 457 2603 179 ...
$ sample_90 : int 631 103 54 55 32 0 0 321 3260 177 ...
$ sample_91 : int 605 315 20 27 0 42 4 396 2186 130 ...
$ sample_92 : int 366 204 27 38 0 34 0 234 2451 134 ...
$ sample_94 : int 339 301 7 60 14 0 0 455 2464 153 ...
$ sample_97 : int 517 56 22 25 0 0 22 279 2768 189 ...
$ sample_98 : int 577 49 63 18 0 26 101 229 1935 282 ...
$ sample_99 : int 235 260 12 50 0 56 0 208 1991 138 ...3.1.2 Convert to matrix
# Convert the data frame to a matrix
gene_count_matrix <- as.matrix(csv_content)
# Print the matrix
print(head(gene_count_matrix)) sample_1 sample_10 sample_100 sample_107
gene-LOC132462341|LOC132462341 360 464 391 346
gene-abce1|abce1 694 325 276 77
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 10 13 11
gene-ube2v1|ube2v1 10 22 15 19
gene-cldn15la|cldn15la 0 9 0 0
gene-muc15|muc15 0 40 40 29
sample_108 sample_109 sample_11 sample_110
gene-LOC132462341|LOC132462341 691 408 436 509
gene-abce1|abce1 409 196 281 284
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 42 14 36
gene-ube2v1|ube2v1 74 28 63 35
gene-cldn15la|cldn15la 18 0 0 0
gene-muc15|muc15 8 0 38 0
sample_116 sample_12 sample_13 sample_18
gene-LOC132462341|LOC132462341 366 373 432 385
gene-abce1|abce1 327 310 393 386
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 9 0
gene-ube2v1|ube2v1 30 31 25 42
gene-cldn15la|cldn15la 0 27 0 0
gene-muc15|muc15 16 15 0 0
sample_19 sample_2 sample_20 sample_21
gene-LOC132462341|LOC132462341 330 288 307 346
gene-abce1|abce1 106 290 252 326
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 3 21
gene-ube2v1|ube2v1 17 14 21 60
gene-cldn15la|cldn15la 0 0 0 0
gene-muc15|muc15 0 0 22 22
sample_28 sample_29 sample_3 sample_30
gene-LOC132462341|LOC132462341 293 347 452 452
gene-abce1|abce1 345 254 363 353
gene-si:dkey-6i22.5|si:dkey-6i22.5 26 9 39 20
gene-ube2v1|ube2v1 61 14 29 37
gene-cldn15la|cldn15la 0 0 4 35
gene-muc15|muc15 69 22 0 15
sample_31 sample_36 sample_4 sample_5
gene-LOC132462341|LOC132462341 984 230 469 400
gene-abce1|abce1 922 160 588 285
gene-si:dkey-6i22.5|si:dkey-6i22.5 10 0 0 10
gene-ube2v1|ube2v1 68 37 0 30
gene-cldn15la|cldn15la 0 0 0 4
gene-muc15|muc15 0 34 55 51
sample_78 sample_79 sample_80 sample_83
gene-LOC132462341|LOC132462341 472 311 368 312
gene-abce1|abce1 178 119 197 134
gene-si:dkey-6i22.5|si:dkey-6i22.5 48 41 60 39
gene-ube2v1|ube2v1 10 13 56 32
gene-cldn15la|cldn15la 0 0 0 0
gene-muc15|muc15 0 30 19 31
sample_88 sample_90 sample_91 sample_92
gene-LOC132462341|LOC132462341 551 631 605 366
gene-abce1|abce1 128 103 315 204
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 54 20 27
gene-ube2v1|ube2v1 45 55 27 38
gene-cldn15la|cldn15la 8 32 0 0
gene-muc15|muc15 0 0 42 34
sample_94 sample_97 sample_98 sample_99
gene-LOC132462341|LOC132462341 339 517 577 235
gene-abce1|abce1 301 56 49 260
gene-si:dkey-6i22.5|si:dkey-6i22.5 7 22 63 12
gene-ube2v1|ube2v1 60 25 18 50
gene-cldn15la|cldn15la 14 0 0 0
gene-muc15|muc15 0 0 26 563.1.3 Load matrix into edgeR
dge <- DGEList(counts = gene_count_matrix, group = factor(temperatures))
dgeAn object of class "DGEList"
$counts
sample_1 sample_10 sample_100 sample_107
gene-LOC132462341|LOC132462341 360 464 391 346
gene-abce1|abce1 694 325 276 77
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 10 13 11
gene-ube2v1|ube2v1 10 22 15 19
gene-cldn15la|cldn15la 0 9 0 0
sample_108 sample_109 sample_11 sample_110
gene-LOC132462341|LOC132462341 691 408 436 509
gene-abce1|abce1 409 196 281 284
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 42 14 36
gene-ube2v1|ube2v1 74 28 63 35
gene-cldn15la|cldn15la 18 0 0 0
sample_116 sample_12 sample_13 sample_18
gene-LOC132462341|LOC132462341 366 373 432 385
gene-abce1|abce1 327 310 393 386
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 9 0
gene-ube2v1|ube2v1 30 31 25 42
gene-cldn15la|cldn15la 0 27 0 0
sample_19 sample_2 sample_20 sample_21
gene-LOC132462341|LOC132462341 330 288 307 346
gene-abce1|abce1 106 290 252 326
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 3 21
gene-ube2v1|ube2v1 17 14 21 60
gene-cldn15la|cldn15la 0 0 0 0
sample_28 sample_29 sample_3 sample_30
gene-LOC132462341|LOC132462341 293 347 452 452
gene-abce1|abce1 345 254 363 353
gene-si:dkey-6i22.5|si:dkey-6i22.5 26 9 39 20
gene-ube2v1|ube2v1 61 14 29 37
gene-cldn15la|cldn15la 0 0 4 35
sample_31 sample_36 sample_4 sample_5
gene-LOC132462341|LOC132462341 984 230 469 400
gene-abce1|abce1 922 160 588 285
gene-si:dkey-6i22.5|si:dkey-6i22.5 10 0 0 10
gene-ube2v1|ube2v1 68 37 0 30
gene-cldn15la|cldn15la 0 0 0 4
sample_78 sample_79 sample_80 sample_83
gene-LOC132462341|LOC132462341 472 311 368 312
gene-abce1|abce1 178 119 197 134
gene-si:dkey-6i22.5|si:dkey-6i22.5 48 41 60 39
gene-ube2v1|ube2v1 10 13 56 32
gene-cldn15la|cldn15la 0 0 0 0
sample_88 sample_90 sample_91 sample_92
gene-LOC132462341|LOC132462341 551 631 605 366
gene-abce1|abce1 128 103 315 204
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 54 20 27
gene-ube2v1|ube2v1 45 55 27 38
gene-cldn15la|cldn15la 8 32 0 0
sample_94 sample_97 sample_98 sample_99
gene-LOC132462341|LOC132462341 339 517 577 235
gene-abce1|abce1 301 56 49 260
gene-si:dkey-6i22.5|si:dkey-6i22.5 7 22 63 12
gene-ube2v1|ube2v1 60 25 18 50
gene-cldn15la|cldn15la 14 0 0 0
30570 more rows ...
$samples
group lib.size norm.factors
sample_1 16C 49994893 1
sample_10 16C 43506227 1
sample_100 9C 44600544 1
sample_107 9C 41991971 1
sample_108 9C 47412580 1
31 more rows ...3.1.4 Filter low counts
Filters for genes with at >= 10 reads across at least 3 samples.
keep <- filterByExpr(dge)
dge <- dge[keep, , keep.lib.sizes=FALSE]
dgeAn object of class "DGEList"
$counts
sample_1 sample_10 sample_100 sample_107
gene-LOC132462341|LOC132462341 360 464 391 346
gene-abce1|abce1 694 325 276 77
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 10 13 11
gene-ube2v1|ube2v1 10 22 15 19
gene-muc15|muc15 0 40 40 29
sample_108 sample_109 sample_11 sample_110
gene-LOC132462341|LOC132462341 691 408 436 509
gene-abce1|abce1 409 196 281 284
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 42 14 36
gene-ube2v1|ube2v1 74 28 63 35
gene-muc15|muc15 8 0 38 0
sample_116 sample_12 sample_13 sample_18
gene-LOC132462341|LOC132462341 366 373 432 385
gene-abce1|abce1 327 310 393 386
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 9 0
gene-ube2v1|ube2v1 30 31 25 42
gene-muc15|muc15 16 15 0 0
sample_19 sample_2 sample_20 sample_21
gene-LOC132462341|LOC132462341 330 288 307 346
gene-abce1|abce1 106 290 252 326
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 3 21
gene-ube2v1|ube2v1 17 14 21 60
gene-muc15|muc15 0 0 22 22
sample_28 sample_29 sample_3 sample_30
gene-LOC132462341|LOC132462341 293 347 452 452
gene-abce1|abce1 345 254 363 353
gene-si:dkey-6i22.5|si:dkey-6i22.5 26 9 39 20
gene-ube2v1|ube2v1 61 14 29 37
gene-muc15|muc15 69 22 0 15
sample_31 sample_36 sample_4 sample_5
gene-LOC132462341|LOC132462341 984 230 469 400
gene-abce1|abce1 922 160 588 285
gene-si:dkey-6i22.5|si:dkey-6i22.5 10 0 0 10
gene-ube2v1|ube2v1 68 37 0 30
gene-muc15|muc15 0 34 55 51
sample_78 sample_79 sample_80 sample_83
gene-LOC132462341|LOC132462341 472 311 368 312
gene-abce1|abce1 178 119 197 134
gene-si:dkey-6i22.5|si:dkey-6i22.5 48 41 60 39
gene-ube2v1|ube2v1 10 13 56 32
gene-muc15|muc15 0 30 19 31
sample_88 sample_90 sample_91 sample_92
gene-LOC132462341|LOC132462341 551 631 605 366
gene-abce1|abce1 128 103 315 204
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 54 20 27
gene-ube2v1|ube2v1 45 55 27 38
gene-muc15|muc15 0 0 42 34
sample_94 sample_97 sample_98 sample_99
gene-LOC132462341|LOC132462341 339 517 577 235
gene-abce1|abce1 301 56 49 260
gene-si:dkey-6i22.5|si:dkey-6i22.5 7 22 63 12
gene-ube2v1|ube2v1 60 25 18 50
gene-muc15|muc15 0 0 26 56
16688 more rows ...
$samples
group lib.size norm.factors
sample_1 16C 49973355 1
sample_10 16C 43459343 1
sample_100 9C 44577844 1
sample_107 9C 41972179 1
sample_108 9C 47313070 1
31 more rows ...3.1.5 Normalization
dge <- calcNormFactors(object = dge)
dgeAn object of class "DGEList"
$counts
sample_1 sample_10 sample_100 sample_107
gene-LOC132462341|LOC132462341 360 464 391 346
gene-abce1|abce1 694 325 276 77
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 10 13 11
gene-ube2v1|ube2v1 10 22 15 19
gene-muc15|muc15 0 40 40 29
sample_108 sample_109 sample_11 sample_110
gene-LOC132462341|LOC132462341 691 408 436 509
gene-abce1|abce1 409 196 281 284
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 42 14 36
gene-ube2v1|ube2v1 74 28 63 35
gene-muc15|muc15 8 0 38 0
sample_116 sample_12 sample_13 sample_18
gene-LOC132462341|LOC132462341 366 373 432 385
gene-abce1|abce1 327 310 393 386
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 9 0
gene-ube2v1|ube2v1 30 31 25 42
gene-muc15|muc15 16 15 0 0
sample_19 sample_2 sample_20 sample_21
gene-LOC132462341|LOC132462341 330 288 307 346
gene-abce1|abce1 106 290 252 326
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 3 21
gene-ube2v1|ube2v1 17 14 21 60
gene-muc15|muc15 0 0 22 22
sample_28 sample_29 sample_3 sample_30
gene-LOC132462341|LOC132462341 293 347 452 452
gene-abce1|abce1 345 254 363 353
gene-si:dkey-6i22.5|si:dkey-6i22.5 26 9 39 20
gene-ube2v1|ube2v1 61 14 29 37
gene-muc15|muc15 69 22 0 15
sample_31 sample_36 sample_4 sample_5
gene-LOC132462341|LOC132462341 984 230 469 400
gene-abce1|abce1 922 160 588 285
gene-si:dkey-6i22.5|si:dkey-6i22.5 10 0 0 10
gene-ube2v1|ube2v1 68 37 0 30
gene-muc15|muc15 0 34 55 51
sample_78 sample_79 sample_80 sample_83
gene-LOC132462341|LOC132462341 472 311 368 312
gene-abce1|abce1 178 119 197 134
gene-si:dkey-6i22.5|si:dkey-6i22.5 48 41 60 39
gene-ube2v1|ube2v1 10 13 56 32
gene-muc15|muc15 0 30 19 31
sample_88 sample_90 sample_91 sample_92
gene-LOC132462341|LOC132462341 551 631 605 366
gene-abce1|abce1 128 103 315 204
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 54 20 27
gene-ube2v1|ube2v1 45 55 27 38
gene-muc15|muc15 0 0 42 34
sample_94 sample_97 sample_98 sample_99
gene-LOC132462341|LOC132462341 339 517 577 235
gene-abce1|abce1 301 56 49 260
gene-si:dkey-6i22.5|si:dkey-6i22.5 7 22 63 12
gene-ube2v1|ube2v1 60 25 18 50
gene-muc15|muc15 0 0 26 56
16688 more rows ...
$samples
group lib.size norm.factors
sample_1 16C 49973355 1.0828322
sample_10 16C 43459343 1.3481915
sample_100 9C 44577844 0.9792758
sample_107 9C 41972179 0.7062689
sample_108 9C 47313070 1.5247621
31 more rows ...3.1.6 Model fitting and estimating dispersions
dge <- estimateDisp(dge)
dgeAn object of class "DGEList"
$counts
sample_1 sample_10 sample_100 sample_107
gene-LOC132462341|LOC132462341 360 464 391 346
gene-abce1|abce1 694 325 276 77
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 10 13 11
gene-ube2v1|ube2v1 10 22 15 19
gene-muc15|muc15 0 40 40 29
sample_108 sample_109 sample_11 sample_110
gene-LOC132462341|LOC132462341 691 408 436 509
gene-abce1|abce1 409 196 281 284
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 42 14 36
gene-ube2v1|ube2v1 74 28 63 35
gene-muc15|muc15 8 0 38 0
sample_116 sample_12 sample_13 sample_18
gene-LOC132462341|LOC132462341 366 373 432 385
gene-abce1|abce1 327 310 393 386
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 9 0
gene-ube2v1|ube2v1 30 31 25 42
gene-muc15|muc15 16 15 0 0
sample_19 sample_2 sample_20 sample_21
gene-LOC132462341|LOC132462341 330 288 307 346
gene-abce1|abce1 106 290 252 326
gene-si:dkey-6i22.5|si:dkey-6i22.5 0 14 3 21
gene-ube2v1|ube2v1 17 14 21 60
gene-muc15|muc15 0 0 22 22
sample_28 sample_29 sample_3 sample_30
gene-LOC132462341|LOC132462341 293 347 452 452
gene-abce1|abce1 345 254 363 353
gene-si:dkey-6i22.5|si:dkey-6i22.5 26 9 39 20
gene-ube2v1|ube2v1 61 14 29 37
gene-muc15|muc15 69 22 0 15
sample_31 sample_36 sample_4 sample_5
gene-LOC132462341|LOC132462341 984 230 469 400
gene-abce1|abce1 922 160 588 285
gene-si:dkey-6i22.5|si:dkey-6i22.5 10 0 0 10
gene-ube2v1|ube2v1 68 37 0 30
gene-muc15|muc15 0 34 55 51
sample_78 sample_79 sample_80 sample_83
gene-LOC132462341|LOC132462341 472 311 368 312
gene-abce1|abce1 178 119 197 134
gene-si:dkey-6i22.5|si:dkey-6i22.5 48 41 60 39
gene-ube2v1|ube2v1 10 13 56 32
gene-muc15|muc15 0 30 19 31
sample_88 sample_90 sample_91 sample_92
gene-LOC132462341|LOC132462341 551 631 605 366
gene-abce1|abce1 128 103 315 204
gene-si:dkey-6i22.5|si:dkey-6i22.5 49 54 20 27
gene-ube2v1|ube2v1 45 55 27 38
gene-muc15|muc15 0 0 42 34
sample_94 sample_97 sample_98 sample_99
gene-LOC132462341|LOC132462341 339 517 577 235
gene-abce1|abce1 301 56 49 260
gene-si:dkey-6i22.5|si:dkey-6i22.5 7 22 63 12
gene-ube2v1|ube2v1 60 25 18 50
gene-muc15|muc15 0 0 26 56
16688 more rows ...
$samples
group lib.size norm.factors
sample_1 16C 49973355 1.0828322
sample_10 16C 43459343 1.3481915
sample_100 9C 44577844 0.9792758
sample_107 9C 41972179 0.7062689
sample_108 9C 47313070 1.5247621
31 more rows ...
$common.dispersion
[1] 0.3258355
$trended.dispersion
[1] 0.1792662 0.2158552 0.8880326 0.7699354 0.9217978
16688 more elements ...
$tagwise.dispersion
[1] 0.1017614 0.1666824 0.9805648 0.3988847 2.8340881
16688 more elements ...
$AveLogCPM
[1] 3.3189831 2.6344211 -0.7839951 -0.2807668 -0.9718511
16688 more elements ...
$trend.method
[1] "locfit"
$prior.df
[1] 4.732584
$prior.n
[1] 0.1391936
$span
[1] 0.29104683.1.7 Exact test (Gene expression)
exact_test_genes <- exactTest(dge)
exact_test_genesAn object of class "DGEExact"
$table
logFC logCPM PValue
gene-LOC132462341|LOC132462341 0.3690768 3.3189831 0.0174459833
gene-abce1|abce1 -0.7406649 2.6344211 0.0002052135
gene-si:dkey-6i22.5|si:dkey-6i22.5 1.8787265 -0.7839951 0.0001945058
gene-ube2v1|ube2v1 0.2838600 -0.2807668 0.3721151448
gene-muc15|muc15 0.0123765 -0.9718511 0.9923746564
16688 more rows ...
$comparison
[1] "16C" "9C"
$genes
NULL3.1.8 Differential gene expression summary
summary(decideTests(object = exact_test_genes, p.value = 0.05)) 9C-16C
Down 2071
NotSig 11980
Up 26423.1.9 Peek at DEGs
Selects all genes (n = "Inf") with adjusted p-value < 0.05, sorts by false discovery rate (adjust.method = "fdr").
Converts object to a data frame (the $table part).
top_degs_table <- topTags(object = exact_test_genes, n = "Inf", adjust.method = "fdr", p.value = 0.05)$table
str(top_degs_table)'data.frame': 16693 obs. of 4 variables:
$ logFC : num 1.9 -2.59 -1.45 -3.13 1.87 ...
$ logCPM: num 6.59 4.06 4.93 3.18 5.57 ...
$ PValue: num 1.26e-64 5.31e-37 2.15e-30 1.48e-26 4.65e-25 ...
$ FDR : num 2.10e-60 4.43e-33 1.19e-26 6.18e-23 1.43e-21 ...3.1.10 Convert gene ID rownames to column
# Step 1: Extract row names and create a new column 'gene_ids'
top_degs_table$gene_ids <- rownames(top_degs_table)
# Step 2: Reorder columns to make 'gene_ids' the first column
top_degs_table <- top_degs_table[, c("gene_ids", names(top_degs_table)[-length(names(top_degs_table))])]
# Step 3: Check the result
head(top_degs_table) gene_ids logFC
gene-LOC132469840|LOC132469840 gene-LOC132469840|LOC132469840 1.897837
gene-serpinh1b|serpinh1b gene-serpinh1b|serpinh1b -2.589704
gene-znf706|znf706 gene-znf706|znf706 -1.450400
gene-hacd4|hacd4 gene-hacd4|hacd4 -3.133038
gene-LOC132448390|LOC132448390 gene-LOC132448390|LOC132448390 1.874161
gene-zgc:103586|zgc:103586 gene-zgc:103586|zgc:103586 1.584728
logCPM PValue FDR
gene-LOC132469840|LOC132469840 6.594858 1.259480e-64 2.102450e-60
gene-serpinh1b|serpinh1b 4.063945 5.307949e-37 4.430280e-33
gene-znf706|znf706 4.929053 2.146565e-30 1.194420e-26
gene-hacd4|hacd4 3.179135 1.480134e-26 6.176968e-23
gene-LOC132448390|LOC132448390 5.570204 4.647165e-25 1.434822e-21
gene-zgc:103586|zgc:103586 5.356047 5.157212e-25 1.434822e-213.1.11 Write DEGs to file
# Write dataframe to CSV
write.csv(
top_degs_table,
file = "../output/13.0.0-RNAseq-edgeR/DEGs_9C-vs-16C-p-0.05.csv",
quote = FALSE,
row.names = FALSE)3.1.12 Volcano plot
ghibli_colors <- ghibli_palette("PonyoMedium", type = "discrete")
ghibli_subset <- c(ghibli_colors[3], ghibli_colors[6], ghibli_colors[4])
# Create new column and fill with `NA`
top_degs_table$topDE <- "NA"
# Set value of "Up" for genes with FC > 1 and FDR < 0.05
top_degs_table$topDE[top_degs_table$logFC > 1 & top_degs_table$FDR < 0.05] <- "Up"
# Set value of "Down" for genes with FC < -1 and FDR < 0.05
top_degs_table$topDE[top_degs_table$logFC < -1 & top_degs_table$FDR < 0.05] <- "Down"
ggplot(data=top_degs_table, aes(x=logFC, y=-log10(FDR), color = topDE)) +
geom_point() +
theme_minimal() +
scale_colour_discrete(type = ghibli_subset, breaks = c("Up", "Down"))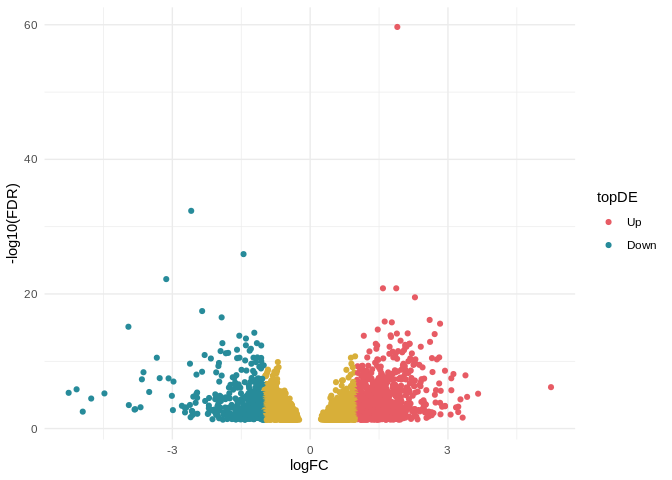
4 CITATIONS
Chen, Yunshun, Lizhong Chen, Aaron T. L. Lun, Pedro L. Baldoni, and Gordon K. Smyth. 2024. “edgeR 4.0: Powerful Differential Analysis of Sequencing Data with Expanded Functionality and Improved Support for Small Counts and Larger Datasets.” http://dx.doi.org/10.1101/2024.01.21.576131.
Chen, Yunshun, Aaron T. L. Lun, and Gordon K. Smyth. 2016. “From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the edgeR Quasi-Likelihood Pipeline.” F1000Research 5 (August): 1438. https://doi.org/10.12688/f1000research.8987.2.
McCarthy, Davis J., Yunshun Chen, and Gordon K. Smyth. 2012. “Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation.” Nucleic Acids Research 40 (10): 4288–97. https://doi.org/10.1093/nar/gks042.
Robinson, Mark D., Davis J. McCarthy, and Gordon K. Smyth. 2009. “edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data.” Bioinformatics 26 (1): 139–40. https://doi.org/10.1093/bioinformatics/btp616.