INTRO
Steven asked that I isolate gDNA from a Pacific cod blood samples (GitHub Issue). As we haven’t isolated gDNA from blood before, Laura Spencer identified a set of samples that could be used for testing (GitHub Issue).
I selected the following four samples for testing:
5B14B65B76B
METHODS
DNA Isolations
DNA was isolated using the Quick-DNA Miniprep Kit (ZymoResearch), according to the manufacturer’s protocol - including the recommended addition of beta-mercaptoethanol to the buffer. Pacific cod blood had been collected and frozen/stored at -80oC, with no preservatives/anticoagulants (e.g. heparin). Blood was thawed on ice and briefly centrifuged (20s @ 10,000g) to collect blood at bottom of tubes. Since the volumes of all blood samples was < 50uL, I added 200uL of g-DNA Lysis Buffer to each tube (per the manufacturer’s protocol for blood isolations). DNA was eventually eluted with 50uL of Elution Buffer.
DNA Quantification
Subsequently, DNA was quantified using the 1x dsDNA Broad Range Qubit Assay (ThermoFisher). One microliter of sample was combined with 199uL of the dye/buffer mix and measured on a Qubit 3.0.
Agarose Gel
After quantification, samples were run on a 0.8% agarose low-TAE gel (with ethidium bromide added) to assess DNA integrity. For the sake of time, I did not load equal quantities of each sample. Instead, I used 0.5uL of DNA from each sample, except 65B. I used 9.5uL of 65B, due to its low concentration (see Qubit Data below). Samples were prepared with a final volume of 12uL;with 2uL of 6X loading dye, and the remainder of volume with water.
The O’Gene Ruler (ThermoFisher) was loaded in Lane 1. The gDNA samples were loaded in numerical order, from left to right in the subsequent lanes.
DNA was stored in the same boxes and positions in which the blood had been stored in the -80oC freezer.
RESULTS
Qubit data
Sample 65B had ridiculously low yield, despite not having noticeably less input sample volume, nor any apparent issues during isolation (e.g. clogged column). It’s not readily apparent why this sample had such low yields.
| sample_ID | Original sample conc.(ng/uL) |
|---|---|
| 5B | 620 |
| 14B | 440 |
| 65B | 5.84 |
| 76B | 862 |
Gel
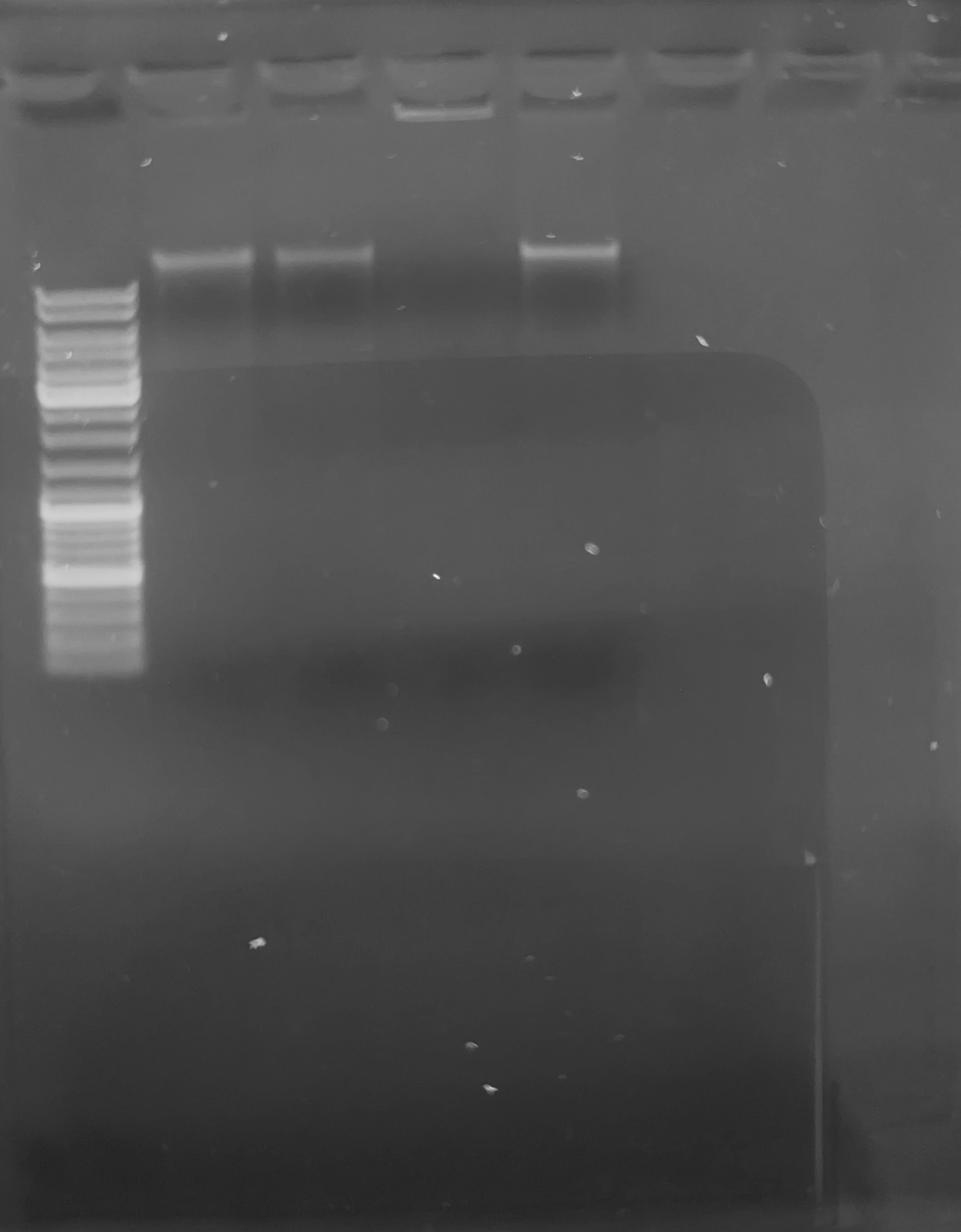
CONCLUSIONS
Overall, it appears that the DNA isolation procedure from direct-frozen Pacific cod blood produces good yields and intact, high-molecular weight DNA. As such, I will proceed with gDNA isolations for the blood samples selected by Laura Spencer (GitHub Issue comment).