INTRO
After previously designing bisulfite PCR primers for the C1Q gene on 20240709, and then screwing up the initial PCR, it was time to redo this the correct way. Due to their length, low complexity, and low melting temperatures (Tm) I opted to test these out on a pool of the Salvelinus namaycush gDNA isolated on 20240712, using a temperature gradient.
MATERIALS & METHODS
DNA Pooling
1uL of each sample was used to create a pool of gDNA.
PCR Reactions
PCR reactions were run using EpiMark Hot Start Taq DNA Polymerase (New England Biolabs), according to the manufacturer’s protocol, in a total reaction volume of 25uL. Since all samples were the same, the equivalent of 1uL of DNA per reaction was added directly to the master mix. The master mix was then distributed to 0.1mL PCR plates and run on a CFX Connect (BioRad) thermalcycler.
I tested three different annealing temps (45oC, 50oC, and 55oC).
PCR Cycling
Annealing temperature gradient PCR was performed in a CFX Connect (BioRad).
Cycling params:
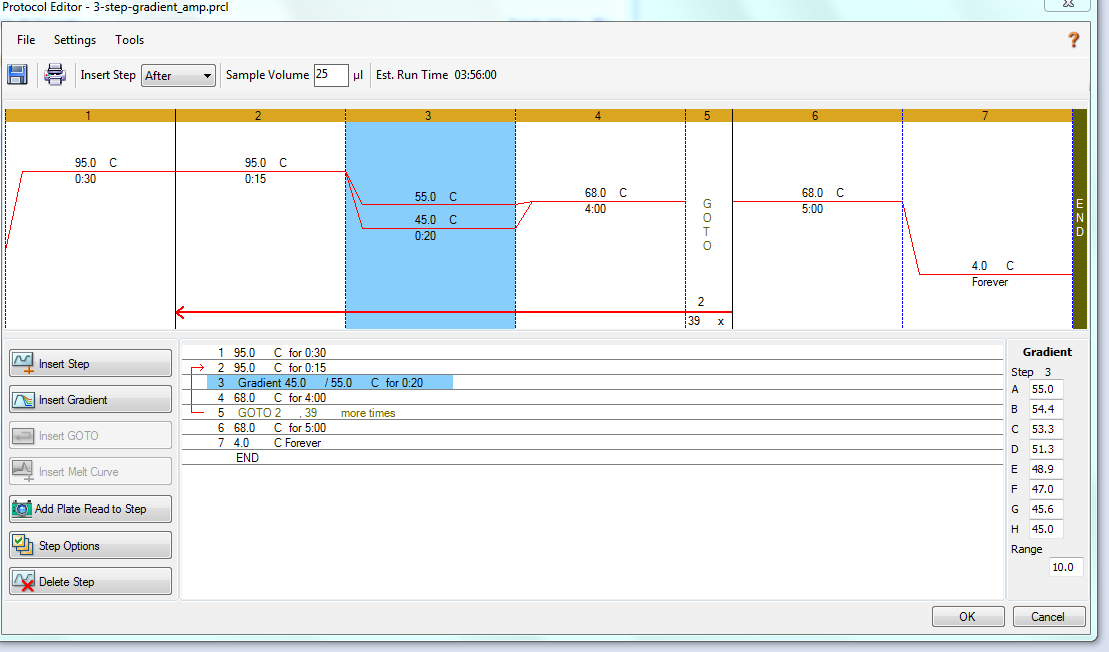
- 95oC - 30s
- 95oC - 15s
- 45 - 55oC - 20s
- 68oC - 4m
- Go to Step 2 x 39.
- 68oC - 5m
- 4oC - FOREVER
Samples were loaded in the following positions:

PCR reactions were loaded on a low-TAE, 1.0% agarose gel, with ethidium bromide. Gel was run at 107V for ~45mins and then imaged.
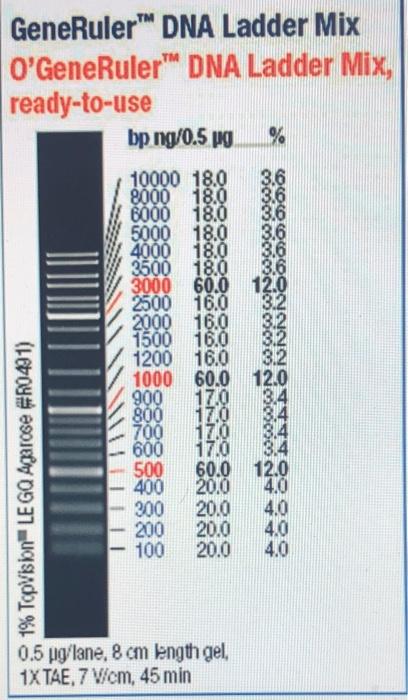
O’GeneRuler (ThermoFisher) Ladder was used (5uL) for size reference:
RESULTS
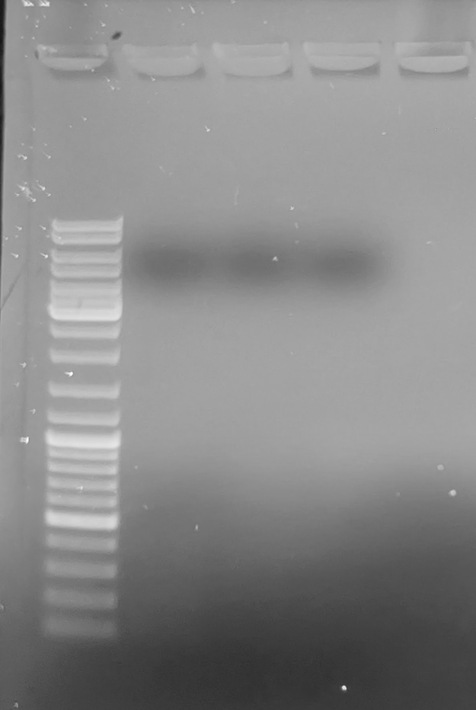
No amplification.
DISCUSSION
Well, that’s a bummer. Will design some regular PCR primers to see if we can succussfully PCR the C1q gene from the original gDNA (i.e. not bisulfite converted DNA).