The following samples were sent out for PacBio sequencing in their entirety on 20241104:
FA047 FA048 FA049 FA051 FA055 FA056
INTRO
Steven asked that I isolate gDNA from a subset of lake trout (S.namaycush) liver samples (GitHub Issue).
METHODS
DNA Isolation
DNA was isolated using the Quick-DNA Miniprep Kit (ZymoResearch), according to the manufacturer’s protocol - including the recommended addition of beta-mercaptoethanol to the lysis buffer. Liver tissues had been stored @ -20oC in RNALater (Ambion). I thawed the tubes, transferred the entire liver tissue (which was very small) to a disposable mortar/pestle combo (1.5mL tube) and homogenized. DNA was eventually eluted with 50uL of Elution Buffer.
Subsequently, DNA was quantified using the 1x dsDNA Broad Range Qubit Assay (ThermoFisher). One microliter of sample was combined with 199uL of the dye/buffer mix and measured on a Qubit 3.0.
DNA was stored in a box labelled “Lake Trout Liver gDNA” in the FTR 209 -20oC freezer.
Agarose gel
Approximately 0.1uL of each sample was run an a 0.8% low-TAE agarose gel, stained with ethidium bromide. This was done just to spot check samples and I wasn’t concerned about loading exact quantities of DNA (or equal quantities) across samples. Sample volumes were mixed with 10uL of H2O and 2uL of 6x Orange DNA Loading Dye (Thermo Scientific).
The gel was run @ 107V for ~45mins and then imaged.
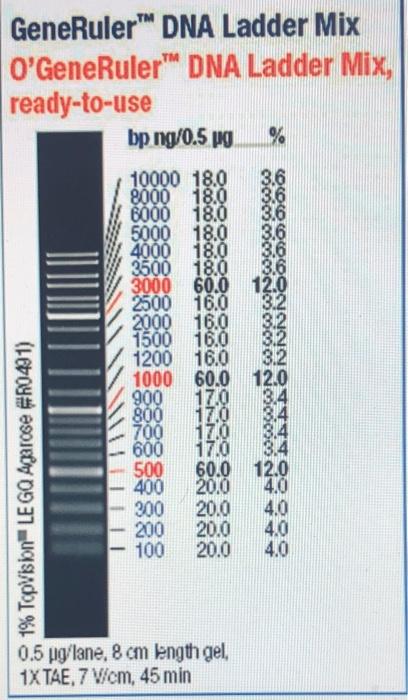
O’GeneRuler DNA Ladder Mix, Ready-to-Use 100-10,00bp (Thermo Scientific) was used (5uL) for size reference:
RESULTS
Qubit data
| sample_ID | Original sample conc. (ng/uL) | yield(ng) |
|---|---|---|
| FA046 | 224 | 8960 |
| FA047 | 984 | 39360 |
| FA048 | 1180 | 47200 |
| FA049 | 584 | 23360 |
| FA050 | 185 | 7400 |
| FA051 | 990 | 39600 |
| FA055 | 770 | 30800 |
| FA056 | 792 | 31680 |
Yield calculation is based on a total sample volume of 40uL. This is likely a conservative volume estimation for each sample, so the total yield is likely a bit higher for each.
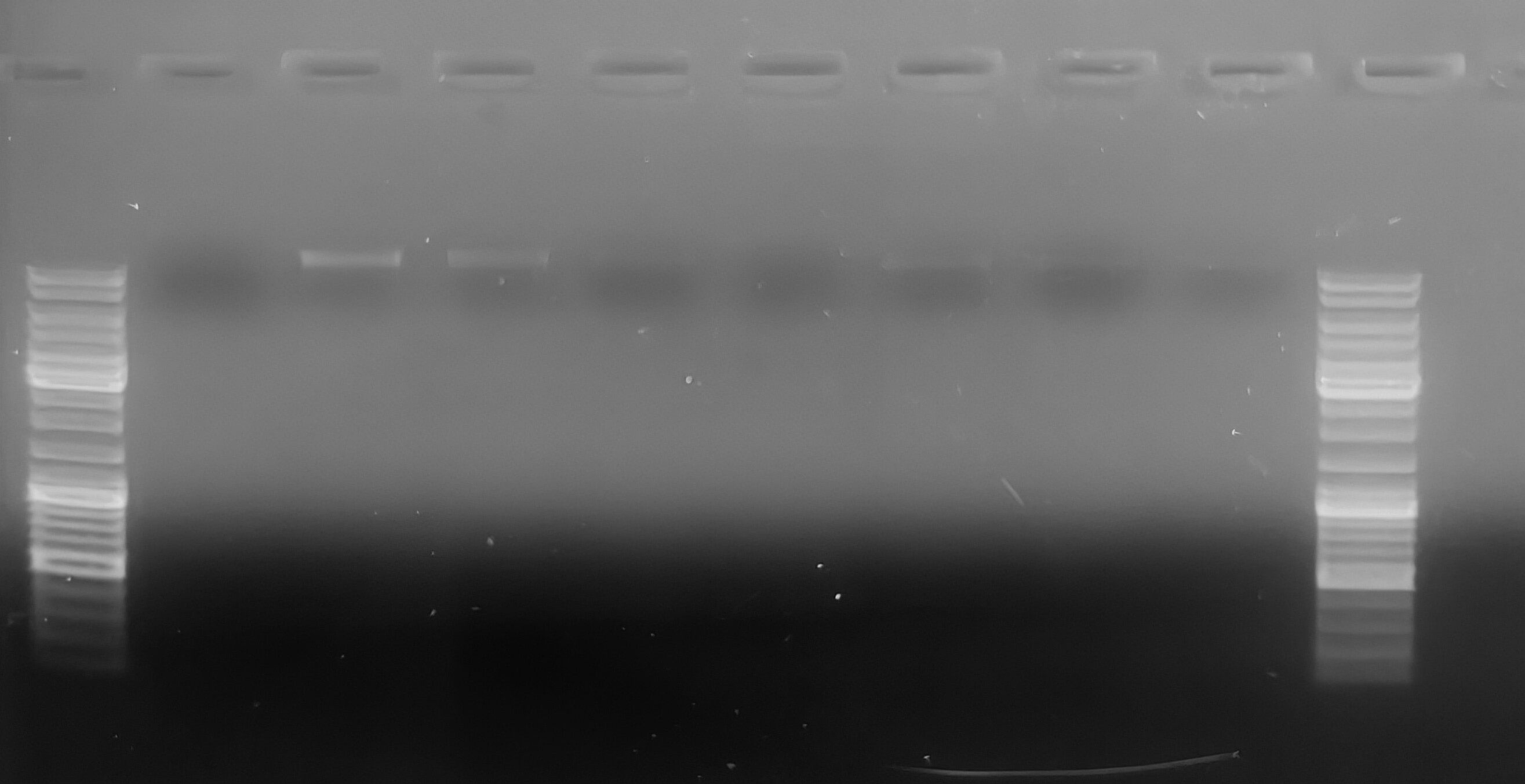
Agarose Gel
Samples were loaded in numerical order, left to right.
Samples FA47 and FA48 have the most prominent bands, but faint bands are visible in all the other lanes (you may need to squint, LOL). Low visibility of bands is most likely due to the inaccuracy involved with attempting to pipette 0.1uL using a P10 pipette… All bands are high molecular weight and don’t show any evidence of degradation (smearing).