INTRO
This notebook is part of the ceasmallr (GitHub repo) project, comparing DNA methylation in Crassostrea virginica (Eastern oyster) larvae and zygotes, via whole-genome bisulfite sequencing (WGBS).
Trimming was done using fastp. After an initial round of trimming, it was discovered that there were problems with some of the FastQ R1/R2 pairs - some did not have an equal number of reads. This leads to an error (and incomplete trimming) in those samples. As such, a step was added to assess these samples and repair them using the BBTools repair.sh script.
Also, trimming had originally been performed on 20220829 (Notebook entry), but had not been fully assessed, which led to the discovery of the trimming errors, and the implemented repair in this notebook.
This notebook was copied from knitted markdown from 00.00-trimming-fastp.Rmd (commit daa2453)
1 Description
This notebook will download raw WGBS FastQs, concatenate them (there were multiple lanes run), and then trim them using fastp. Samples which generate an error during trimming will attempt to be repaired using BBTools repairl.sh (BBMap – Bushnell B. – sourceforge.net/projects/bbmap/). Trimming results will be analyzed with FastQC and summarized by MultiQC (Ewels et al. 2016).
1.1 Inputs
Raw FastQ files with the following pattern:
*.fastq.gz
1.2 Outputs
The expected outputs will be:
multiqc_report.html: A summary report of the alignment results generated by MultiQC, in HTML format.*fastp-trim*.fq.gz: Trimmed FastQ files.Due to the large file sizes of FastQs, these cannot be hosted in the ceasmallr GitHub repo. As such these files are available for download here:
2 Create a Bash variables file
This allows usage of Bash variables across R Markdown chunks.
{
echo "#### Assign Variables ####"
echo ""
echo "# Data directories"
echo 'export repo_dir=/home/shared/8TB_HDD_01/sam/gitrepos/ceasmallr'
echo 'export output_dir_top=${repo_dir}/output/00.00-trimming-fastp'
echo 'export raw_reads_url="https://owl.fish.washington.edu/nightingales/C_virginica/2018OALarvae_DNAm_discovery/"'
echo 'export raw_reads_dir="${repo_dir}/data/raw_reads"'
echo 'export trimmed_fastqs_dir="${output_dir_top}/trimmed-fastqs"'
echo ""
echo "# Paths to programs"
echo 'export programs_dir="/home/shared"'
echo 'export bbmap_repair="${programs_dir}/bbmap-39.12/repair.sh"'
echo 'export bismark_dir="${programs_dir}/Bismark-0.24.0"'
echo 'export bowtie2_dir="${programs_dir}/bowtie2-2.4.4-linux-x86_64"'
echo 'export fastp="${programs_dir}/fastp-v0.24.0/fastp"'
echo 'export fastqc="${programs_dir}/FastQC-0.12.1/fastqc"'
echo 'export multiqc="/home/sam/programs/mambaforge/bin/multiqc"'
echo 'export samtools_dir="${programs_dir}/samtools-1.12"'
echo ""
echo "# Set FastQ filename patterns"
echo "export fastq_pattern='*.fastq.gz'"
echo "export R1_fastq_pattern='*_R1_*.fastq.gz'"
echo "export R2_fastq_pattern='*_R2_*.fastq.gz'"
echo "export trimmed_fastq_pattern='*fastp-trim*.fq.gz'"
echo ""
echo "# Set number of CPUs to use"
echo 'export threads=40'
echo ""
echo "## Inititalize arrays"
echo 'export fastq_array_R1=()'
echo 'export fastq_array_R2=()'
echo 'export trimmed_fastqs_array=()'
echo 'export R1_names_array=()'
echo 'export R2_names_array=()'
echo ""
echo "# Print formatting"
echo 'export line="--------------------------------------------------------"'
echo ""
} > .bashvars
cat .bashvars#### Assign Variables ####
# Data directories
export repo_dir=/home/shared/8TB_HDD_01/sam/gitrepos/ceasmallr
export output_dir_top=${repo_dir}/output/00.00-trimming-fastp
export raw_reads_url="https://owl.fish.washington.edu/nightingales/C_virginica/2018OALarvae_DNAm_discovery/"
export raw_reads_dir="${repo_dir}/data/raw_reads"
export trimmed_fastqs_dir="${output_dir_top}/trimmed-fastqs"
# Paths to programs
export programs_dir="/home/shared"
export bbmap_repair="${programs_dir}/bbmap-39.12/repair.sh"
export bismark_dir="${programs_dir}/Bismark-0.24.0"
export bowtie2_dir="${programs_dir}/bowtie2-2.4.4-linux-x86_64"
export fastp="${programs_dir}/fastp-v0.24.0/fastp"
export fastqc="${programs_dir}/FastQC-0.12.1/fastqc"
export multiqc="/home/sam/programs/mambaforge/bin/multiqc"
export samtools_dir="${programs_dir}/samtools-1.12"
# Set FastQ filename patterns
export fastq_pattern='*.fastq.gz'
export R1_fastq_pattern='*_R1_*.fastq.gz'
export R2_fastq_pattern='*_R2_*.fastq.gz'
export trimmed_fastq_pattern='*fastp-trim*.fq.gz'
# Set number of CPUs to use
export threads=40
## Inititalize arrays
export fastq_array_R1=()
export fastq_array_R2=()
export trimmed_fastqs_array=()
export R1_names_array=()
export R2_names_array=()
# Print formatting
export line="--------------------------------------------------------"3 Download raw reads
The --cut-dirs 2 command cuts the preceding directory structure (i.e. nightingales/C_virginica/) so that we just end up with the reads.
# Load bash variables into memory
source .bashvars
# Create directory, if it doesn't exist
mkdir --parents ${raw_reads_dir}
wget \
--directory-prefix ${raw_reads_dir} \
--recursive \
--no-check-certificate \
--continue \
--cut-dirs 2 \
--no-parent \
--no-host-directories \
--quiet \
${raw_reads_url}
ls -lh "${raw_reads_dir}"3.1 Verify checkums
# Load bash variables into memory
source .bashvars
cd "${raw_reads_dir}"/2018OALarvae_DNAm_discovery
md5sum --check md5sum_list.txt
echo ""
echo "${line}"
echo ""
cd second_lane
md5sum --check md5sum_list.txt4 Concatenate reads
Concatenation also handles samples where there might be a missing set of R2 reads in the second round of sequencing.
# Load bash variables into memory
source .bashvars
cd "${raw_reads_dir}"
# Concatenate FastQ files from 1st and 2nd runs
# Do NOT quote fastq_pattern variable
# Declare an associative array to keep track of processed files
declare -A processed_files
for first_run_fastq in "${raw_reads_dir}"/2018OALarvae_DNAm_discovery/${fastq_pattern}
do
# Strip full path to just get filename.
first_run_fastq_name="${first_run_fastq##*/}"
# Initialize a flag to check if a match is found
match_found=false
# Process second run and concatenate with corresponding FastQ from first run
# Do NOT quote fastq_pattern variable
for second_run_fastq in "${raw_reads_dir}"/2018OALarvae_DNAm_discovery/second_lane/${fastq_pattern}
do
# Strip full path to just get filename.
second_run_fastq_name="${second_run_fastq##*/}"
# Concatenate FastQs with same filenames
if [[ "${first_run_fastq_name}" == "${second_run_fastq_name}" ]]
then
echo "Concatenating ${first_run_fastq} with ${second_run_fastq} to ${raw_reads_dir}/${first_run_fastq_name}"
echo ""
cat "${first_run_fastq}" "${second_run_fastq}" >> "${raw_reads_dir}/${first_run_fastq_name}"
match_found=true
processed_files["${first_run_fastq_name}"]=true
break
fi
done
# If no match is found, copy the file to the target directory
if [[ "${match_found}" == false ]]
then
if [[ -z "${processed_files[${first_run_fastq_name}]}" ]]
then
echo "NO MATCH!"
echo "Copying ${first_run_fastq} to ${raw_reads_dir}"
echo ""
cp "${first_run_fastq}" "${raw_reads_dir}"
processed_files["${first_run_fastq_name}"]=true
fi
fi
echo "Generating checksums for concatenated FastQs..."
md5sum "${first_run_fastq_name}" | tee --append "${first_run_fastq_name}".md5
echo ""
echo "${line}"
echo ""
done5 Trimming and Error Repair
Trimming will remove any Illumina sequencing adapters, as well as polyG and polyX (primarily polyA) sequences. Trimming will also remove the first 15bp from the 5’ end of each read.
Samples generating an error during trimming will attempt to be repaired with BBTools’ repair.sh script.
# Load bash variables into memory
source .bashvars
# Make output directory, if it doesn't exist
mkdir --parents ${output_dir_top}
cd "${raw_reads_dir}"
# Create arrays of fastq R1 files and sample names
# Do NOT quote R1_fastq_pattern variable
for fastq in ${R1_fastq_pattern}
do
fastq_array_R1+=("${fastq}")
# Use parameter substitution to remove all text up to and including last "." from
# right side of string.
R1_names_array+=("${fastq%%.*}")
done
# Create array of fastq R2 files
# Do NOT quote R2_fastq_pattern variable
for fastq in ${R2_fastq_pattern}
do
fastq_array_R2+=("${fastq}")
# Use parameter substitution to remove all text up to and including last "." from
# right side of string.
R2_names_array+=(${fastq%%.*})
done
# Run fastp on files
# Adds JSON report output for downstream usage by MultiQC
echo "Beginning fastp trimming."
echo ""
for index in "${!fastq_array_R1[@]}"
do
R1_sample_name="${R1_names_array[index]}"
R2_sample_name="${R2_names_array[index]}"
stderr_PE_name=$(echo ${R1_sample_name} | awk -F"_" '{print $1}')
${fastp} \
--in1 ${fastq_array_R1[index]} \
--in2 ${fastq_array_R2[index]} \
--detect_adapter_for_pe \
--trim_poly_g \
--trim_poly_x \
--thread 16 \
--trim_front1 15 \
--trim_front2 15 \
--html ${output_dir_top}/"${R1_sample_name}".fastp-trim.report.html \
--json ${output_dir_top}/"${R1_sample_name}".fastp-trim.report.json \
--out1 ${output_dir_top}/"${R1_sample_name}".fastp-trim.fq.gz \
--out2 ${output_dir_top}/"${R2_sample_name}".fastp-trim.fq.gz \
2> ${output_dir_top}/"${stderr_PE_name}".fastp-trim.stderr
grep --before-context 5 "ERROR" ${output_dir_top}/"${stderr_PE_name}".fastp-trim.stderr
# Check for fastp errors and then repair
if grep --quiet "ERROR" ${output_dir_top}/"${stderr_PE_name}".fastp-trim.stderr; then
rm ${output_dir_top}/"${R1_sample_name}".fastp-trim.fq.gz
rm ${output_dir_top}/"${R2_sample_name}".fastp-trim.fq.gz
${bbmap_repair} \
in1=${fastq_array_R1[index]} \
in2=${fastq_array_R2[index]} \
out1="${R1_sample_name}".REPAIRED.fastq.gz \
out2="${R2_sample_name}".REPAIRED.fastq.gz \
outs=/dev/null \
2> "${R1_sample_name}".REPAIRED.stderr
${fastp} \
--in1 "${R1_sample_name}".REPAIRED.fastq.gz \
--in2 "${R2_sample_name}".REPAIRED.fastq.gz \
--detect_adapter_for_pe \
--trim_poly_g \
--trim_poly_x \
--thread 16 \
--trim_front1 15 \
--trim_front2 15 \
--html ${output_dir_top}/"${R1_sample_name}".fastp-trim.REPAIRED.report.html \
--json ${output_dir_top}/"${R1_sample_name}".fastp-trim.REPAIRED.report.json \
--out1 ${output_dir_top}/"${R1_sample_name}".fastp-trim.REPAIRED.fq.gz \
--out2 ${output_dir_top}/"${R2_sample_name}".fastp-trim.REPAIRED.fq.gz \
2> ${output_dir_top}/"${stderr_PE_name}".fastp-trim.REPAIRED.stderr
if grep --quiet "ERROR" ${output_dir_top}/"${stderr_PE_name}".fastp-trim.REPAIRED.stderr; then
echo "These ${stderr_PE_name} samples are broken."
echo "Just give up. :("
echo ""
fi
fi
echo "Finished trimming:"
echo "${fastq_array_R1[index]}"
echo "${fastq_array_R1[index]}"
echo ""
# Generate md5 checksums for newly trimmed files
cd "${output_dir_top}"
md5sum "${R1_sample_name}".fastp-trim.fq.gz > "${R1_sample_name}".fastp-trim.fq.gz.md5
md5sum "${R2_sample_name}".fastp-trim.fq.gz > "${R2_sample_name}".fastp-trim.fq.gz.md5
cd "${raw_reads_dir}"
done6 FastQC
# Load bash variables into memory
source .bashvars
cd "${output_dir_top}"
############ RUN FASTQC ############
# Create array of trimmed FastQs
trimmed_fastqs_array=(${trimmed_fastq_pattern})
# Pass array contents to new variable as space-delimited list
trimmed_fastqc_list=$(echo "${trimmed_fastqs_array[*]}")
echo "Beginning FastQC on trimmed reads..."
echo ""
# Run FastQC
### NOTE: Do NOT quote raw_fastqc_list
${fastqc} \
--threads ${threads} \
--outdir "${output_dir_top}" \
--quiet \
${trimmed_fastqc_list}
echo "FastQC on trimmed reads complete!"
echo ""
############ END FASTQC ############7 MultiQC
# Load bash variables into memory
source .bashvars
cd "${output_dir_top}"
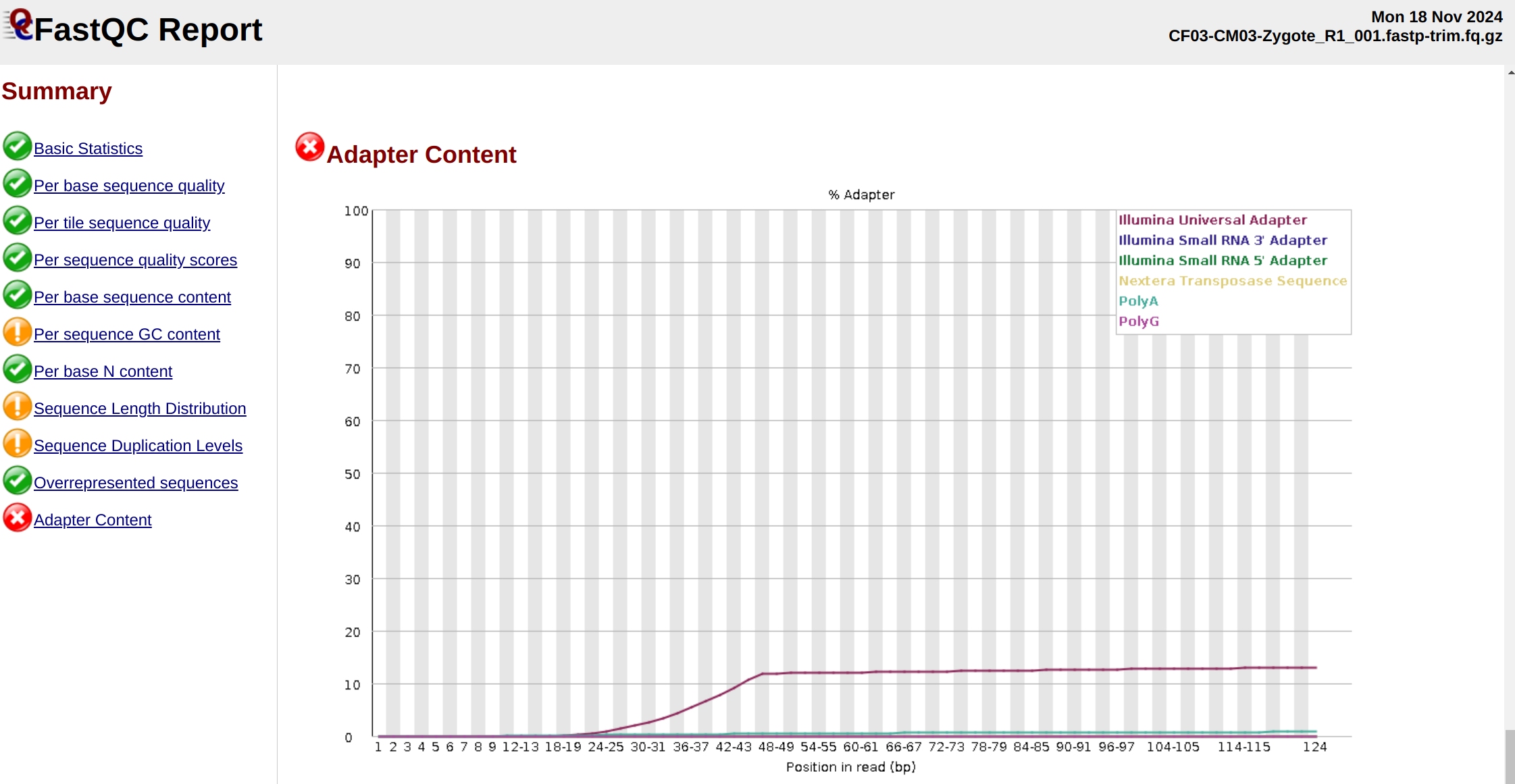
${multiqc} .There are two files which have excessive polyG sequences after trimming, despite specifying fastp to trim polyG sequences
CF03-CM03-Zygote_R1EF04-EM04-Zygote_R1
Each of the corresponging R2 samples has excessive polyA sequences present.