INTRO
Continuing work with project-gigas-carryover lifestage_carryover (GitHub repo) after creating cDNA on 20241024 (Notebook entry), it was finally time to run the qPCRs. Three primer sets were run on 20241210 (Notebook entry): GAPDH, ATP synthase, and cGAS. Four primer sets were run yesterday, 20241211 (Notebook entry): citrate synthase, DNMT1, HSP70 and HSP90.
This notebook describes how the qPCRs were run and links to the various output files. It also provides a brief overview of each primer set’s amplification profiles. This notebook does not have any analysis. This will be performed later.
Due to unequal quantities of RNA used to make cDNA, information from Cq values will not provide meaningful information without normalization first.
| SR ID | Primer Name |
|---|---|
| 1829 | Cg_VIPERIN_F |
| 1828 | Cg_VIPERIN_R |
All samples were run in triplicate, on low-profile, white 96-well plates (USA Scientific) in a CFX Connect (Bio-Rad) or CFX96 (Bio-Rad) real-time thermalcycler. All reactions consisted of the following:
| Component | Stock Concentration | Volume (uL) |
|---|---|---|
| cDNA | NA | 1 |
| SsoAdvanced Universal SYBR Green Supermix (BioRad) | 2x | 10 |
| PF | 10uM | 0.5 |
| PR | 10uM | 0.5 |
| H2O | NA | 8 |
| TOTAL | 20 |
Master mixes were distributed across 2.5 plates for each primer set. No template controls (NTC) were loaded on on just one of the three plates (plate #3 in all instances).
For cycling parameters, plate layouts, etc. see the RESULTS section below.
RESULTS
Summary
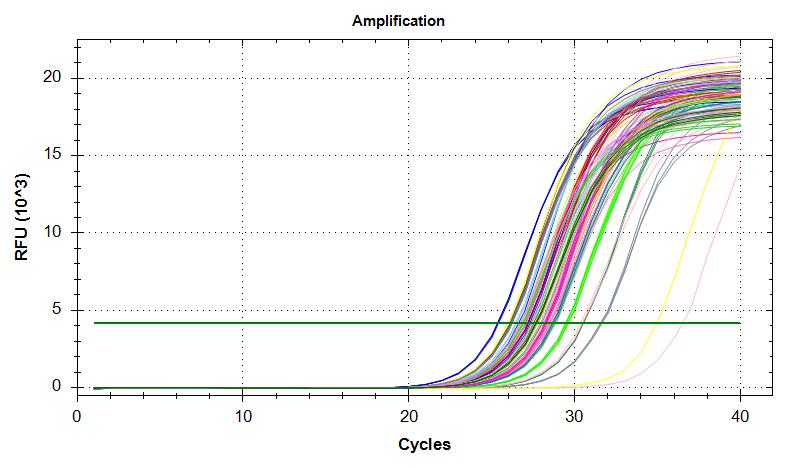
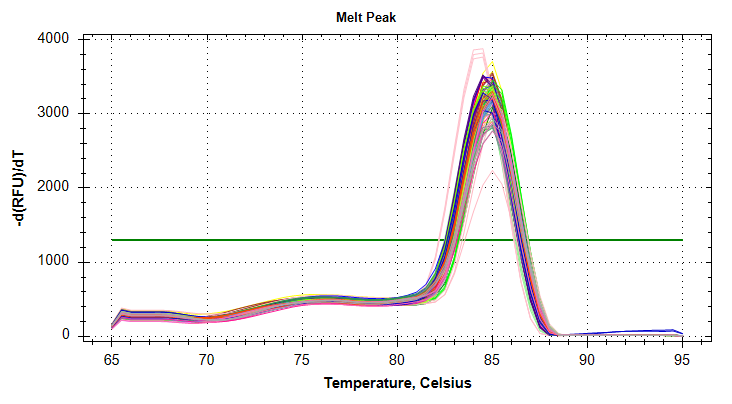
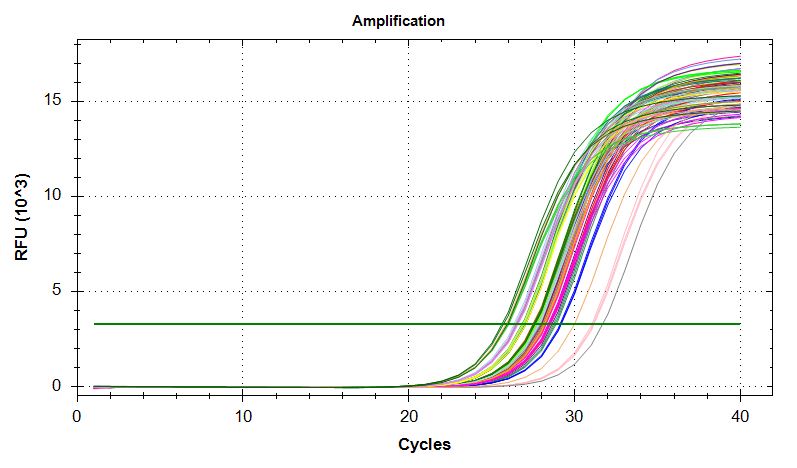
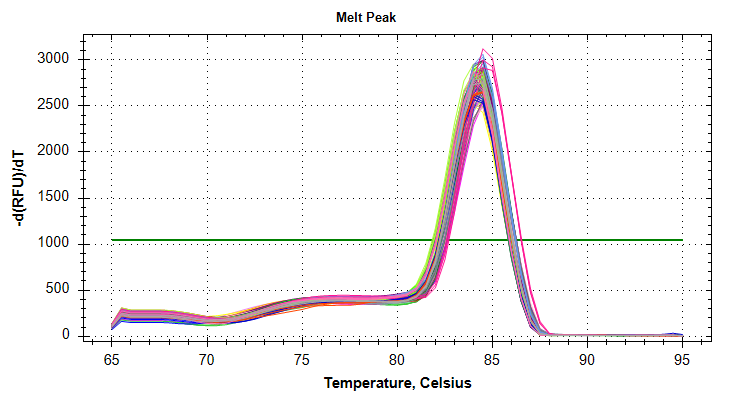
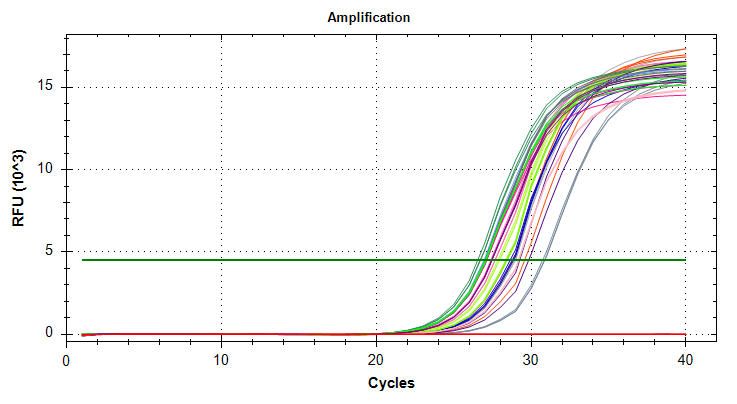
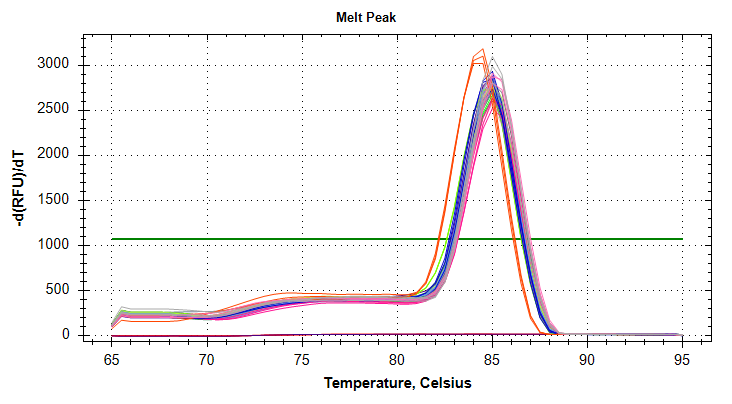
VIPERIN: Amplification and melt plots look good. No amplification in NTCs.
Files
*.pdf: qPCR Reports. Contains plate layouts, cycling params, amp/melt plots, etc.*Amplification-Results_SYBR.csv: Raw fluorescence data.*Cq-Results.csv: Cycle quantity (Cq) data.*.pcrd: Source qPCR data file. Requires CFX Maestro (Bio-Rad) software to open.
All files linked below are from commit e816421.
VIPERIN
Cq Data
Reports
Raw fluorescence
CFX Files
PLOTS