INTRO
This notebook annotated expressed genes with gene ontologies, assigned them to GOslims, and generated corresponding counts of GOs in each GOslim category, as part of urol-e5/deep-dive-expression (GitHub repo).
The contents below are from markdown knitted from 30.00-Apul-transcriptome-GOslims.md (commit 69ad137).
1 BACKGROUND
This notebook will perform annotation of expressed genes, as previously determined by 07-Apul-Hisat.qmd (GitHub).
Briefly, the notebook will perform the following tasks:
Extract all genes from the genome, as GFF and FastA.
Create a subset of only expressed genes, based on gene count matrix.
BLASTx expressed genes against SwissProt database.
Get gene ontology.
Map gene ontology to GOslims and get counts.
Expressed genes were defined as those genes having at least one count across all samples.
1.1 INPUTS
Gene count matrix
Genome FastA
Genome GFF
1.2 OUTPUTS
Genes BED
Genes FastA
Expressed genes FastA
Expressed genes SwissProt IDs only file.
Expressed genes to SwissProt IDs mapping file.
Expressed genes to SwissProtIDs and GO mapping file.
Counts file of expressed genes GOslims.
1.3 SOFTWARE
DIAMOND BLAST (Buchfink, Reuter, and Drost 2021)
bedtools (Quinlan and Hall 2010)
samtools (Danecek et al. 2021)
Biostrings (H. Pagès 2017) (Bioconductor R package)
GO.db (Carlson 2017) (Bioconductor R package)
GSEABase (Martin Morgan 2017) (Bioconductor R package)
2 VARIABLES
# PROGRAMS
blastdbs_dir <- file.path("..", "..", "M-multi-species", "data", "blastdbs")
programs_dir <- file.path("", "home", "shared")
bedtools_dir <- file.path(programs_dir, "bedtools-v2.30.0", "bin")
blast_dir <- file.path(programs_dir, "ncbi-blast-2.15.0+", "bin")
diamond <- file.path(programs_dir, "diamond-2.1.8")
fastaFromBed <- file.path(bedtools_dir, "fastaFromBed")
samtools_dir <- file.path(programs_dir, "samtools-1.12")
samtools <- file.path(samtools_dir, "samtools")
# FILES
count_matrix <- "../output/07-Apul-Hisat/Apul-gene_count_matrix.csv"
diamond_db <- "20250618-diamond"
diamond_output <- file.path("..", "output", "30.00-Apul-transcriptome-GOslims", "Apulchra-expressed-genes.blastx.outfmt6")
genome_fasta <- file.path("..", "data", "Apulcra-genome.fa")
genes_fasta <- file.path("..", "output", "30.00-Apul-transcriptome-GOslims", "Apulchra-genes.fasta")
genes_fasta_index <- file.path("..", "output", "30.00-Apul-transcriptome-GOslims", "Apulchra-genes.fasta.fai")
genes_subset_fasta <- file.path("..", "output", "30.00-Apul-transcriptome-GOslims", "Apulchra-subset-genes.fasta")
genes_subset_fasta_index <- file.path("..", "output", "30.00-Apul-transcriptome-GOslims", "Apulchra-subset-genes.fasta.fai")
genes_bed <- file.path("..", "output", "30.00-Apul-transcriptome-GOslims", "Apulchra-genes.bed")
og_genome_gff <- file.path("..", "data", "Apulcra-genome.gff")
# THREADS
threads <- "40"
##### Official GO info - no need to change #####
goslims_obo <- "goslim_generic.obo"
goslims_url <- "http://current.geneontology.org/ontology/subsets/goslim_generic.obo"
# FORMATTING
line <- "-----------------------------------------------"
# Export these as environment variables for bash chunks.
Sys.setenv(
blastdbs_dir = blastdbs_dir,
count_matrix = count_matrix,
diamond = diamond,
diamond_db = diamond_db,
diamond_output = diamond_output,
fastaFromBed = fastaFromBed,
genes_bed = genes_bed,
genes_fasta = genes_fasta,
genes_fasta_index = genes_fasta_index,
genes_subset_fasta = genes_subset_fasta,
genes_subset_fasta_index = genes_subset_fasta_index,
genome_fasta = genome_fasta,
og_genome_gff = og_genome_gff,
line = line,
samtools = samtools,
threads = threads
)3 Extract genes as FastA
3.1 Extract genes from GFF
# Read GFF, skipping comment lines
genome_gff <- readr::read_tsv(
og_genome_gff,
comment = "#",
col_names = c("seqid", "source", "type", "start", "end", "score", "strand", "phase", "attributes")
)
# Filter for gene features
genes <- as.data.frame(genome_gff) %>%
filter(type == "gene") %>%
mutate(
chrom = seqid,
start = start - 1, # BED is 0-based
end = end,
gene_id = sub("ID=([^;]+);?.*", "\\1", attributes)
) %>%
dplyr::select(chrom, start, end, gene_id)
str(genes)'data.frame': 44371 obs. of 4 variables:
$ chrom : chr "ntLink_0" "ntLink_0" "ntLink_0" "ntLink_0" ...
$ start : num 1104 10214 32056 34823 45952 ...
$ end : num 7056 15286 33275 42794 51024 ...
$ gene_id: chr "FUN_000001" "FUN_000002" "FUN_000003" "FUN_000004" ...3.1.1 Write genes to BED
# Write to BED
write_tsv(genes, genes_bed, col_names = FALSE)3.2 Create genes FastA
"${fastaFromBed}" -fi "${genome_fasta}" -bed "${genes_bed}" -nameOnly > "${genes_fasta}"
# Create FastA index
"${samtools}" faidx "${genes_fasta}"
head "${genes_fasta}"
echo ""
echo ""
head "${genes_fasta_index}">FUN_000001
ATGATGCCACAAGGGTACAAAAACGCCCTTCCAGGCTATAAAGACCTCTATCTTAGCCAAGCAATCACCGAGGAGGTTCACAATGTAAGTTTGTCCTTTTAGCTTGTGCCTGTTtttcctagtttgtttgcttgtttctttctttctttctttctttttagttctattttgtttcattTGTGATTCTTCAAGATATTTGGTGGTTACTTGTTCAGTCTAGCGTAGCCAAACTATTTGAACACTACAAGATAATTCGCAAGAAACAGGTTGAAGAAAATTCCATTTCAAGCGAAATCTTATTTCATTTTTTTACATTCTCTGTAGCTAATATACTGGCAAGTAATTTTATCGCCAACTGAAGGCAAGCACCAAGGACGTTTACATGGTAAGTTTGTCCTTTTGTTACACAAATTGGGTAGTCTGTTTCCTGGTGTTTTTTTTTTTTTTAATGTTTTAATTCACGATTATTCAGGATATCTAGGGATGAATTATCTGTTTATTACTCCAACATAATTAGTAAGAAACCAATTGGGAAGAATTCCAATAGAAGCAAAATCTTAATCTGTCTTTAAACTACTATGTAGTTAATACTTTAGCGAGTGATCTCTTCTCGGTGGCTATTACCATTTGTTGTGAATGCTATGAACGAAGAGCACCTTATTTTACTACCCGACATTATTGTAGCCCTGAGTTCCCTTTAGATCTGACGCTGCCATTTCTTACTTTAAGGCCTCTTCAATTTCTTGGTATTCGTAGATGTTTTCCACTGGCATTGACTGTGGTGTAAGTTCTTTCAAGCACGGGCCGTCGTTGTCTCTTCTAGCACTAGACAAAAAGGTGAGTGAGTTAAAACATGTTGGCTGTTAGTTCTTTCTACTATTTTCAGTTTTGCACTTGAATTCGTTGGAAGCCTCTGTGACAAGGCTTGATTTAATACTCGCTTTTTGGCATTTCAATTTCGCTTTCACCAAGAGAAACTCGATATTTCGTTGACTTCGCAATAAGTTAGTAGTTTTTCTTTCATATTGAGAGCTTCGCATTAAGGGTTCACATCGATGCTCTTGACTAGGCAAAGAAACGAGTCATTGTGCGAATTGCAGCATCTGTTTCCTACAATCACATATCTACACATATTTCTGGTGGTTTTAATTATAGTATCAATCAATCATCTTGGTTGAATATTGGTTAGGCAAAAGAGCATCCCAAAAAAACGTTTCGATCGAGCGTGTGACCAAACGTGTGGTATGACGGGATTCCTGGTATTGCAATGACCACTGATTTAACACATGAAATGGATACCTTAGAAATATTTCCTGTGATAGTGGTTTTTAAGACATTGTGCTAAATGCCACCCTGGATACTTTATCAACAGCAATTTTGGCCTCAGTAGGCACGTTTAGTAACCTCGAATTAACGCCACTTAATTTTTTTGGCAGCTCTGTGTTATCATATTCAAGTTATTACTCCAAAGCCACCTTCCCCTGTTTTGTCTGCGATCGTTGAACACAACGGTGAGTGAGAACTGAATATCAATATTACTATTAATATATCTTGCTTATAGCAGTAATTGAATTTGCTGCGGATGTACAACAATACTTATATTGTTGTTTTGCACCTCAATTAAGCTCAATTTAACGTCACTTAACTTTCTTGGCAGCTCTGTGTCATCGCATTGATCGAGTCATTATTCCAAGTACGTGGCCTTCCCTTCTGCGCACGATCGCTGAACAAAACGGTGAGTGAGAAAACTGAAGCGTTACATGGAAACTAACTGTAGTCAAGTCGCATCAAGGTTTCACCCTTCTAGAACGACTCAGTTAGCTGTCGTTTTGCTTCAAGTTTTTTTCCGGCAGTACTTAGTAGCATAATTTTGACCATTGACCGCTAATTATTGTGCTCAAAATCTCTCCGCAGATTTCTTTGGTGCGTTTGGCTGGGGCATTTCACCGTAAACACGAGCCCCCTTTGACTCCGTGTCCTCGAACGTCGCAAGAAACAGTGAGTAAAACACTTTGTCTGTATGATTCAACTCCATCAGCTGCATTTGCATGTCAGAAAAGCGTTAATTATCGTTTCATTTCCCTTTTATCTACTTTCAATAACTTGTTTCTTTAGAAAAGAAATTTTATATTCGATAGGCTATCTCACTATTTAGAAAAGAGACAACTTACCTTTGGCAAGCAATTTGATTTTCACTCCTATCATTCAATTGATCATGCCATGTTAAGTATTATTCATAAAATCAATGACCGCTAATTATTATGCTCAAAATCTCCCCGCAGATTTCTTTAGTGCATTTGGACAGGGCGCCGTTTCACCGTAAACACGAGCCCCCTTTGATTCCGTGTCCTCGAACGTCGCAAGAAACGGTGAGTAAAACACTTTGTCTATATGATTCAACTCCATCAGTTGCATTAAATATGTCAGAAAAGCGTTAATTATCGTTTCATTTCCCTTTTATCTTTTTTCAATAACTTGTTACTTTAGAAAAGAAATTTGCATTCGATAGGCTGTCTGACTATTTAGAAAAGAGACAACTTACCTTTGGCAAGCAATTTGATTTTCACTCCTATCATTCAATTGATCATGCCATGTTAAGTATTATTCATAAAATCAATGACCGCTAATTATTATGCTCAAATTCTTCCCGCAGATTTCTTTAGTGCATTTGGACAGGGCGCCGTTTCACCGTAAACACGAGCCCCCTTTGACTCCGTGTCCTCGAACGTCGCAAGAAACGGTGAGTAAAACACTTTGTCTATATGATTTAACTCCATCAGCTGCATTTATATGTTAGAAAAGCTTTAATTATCGTTTCATTTTCCCTTTTATCTATTTTCAATAAGTTGTTGCTGTAGAAAAAAAAAAACTTGTATTCAATAGGCTATCTGACTATTTAGAAAAGAGTCAAGTTACCTTTAGTAAGCAATTTGATTTTTGCACCCATGCAACATTCATCTGATCATGCCAAGTTAAGTATTGGTGATCAACTGCAAAAGGCAATTGCTGGAGATTTTTTAGGCTTGAGCAAAGCTTATTTACCTAGAAAGAAGTCACGTTCTTCCTCACACTGAAAACATGCCAACTTTCTTTCTTTTTCTGACAAGAACGAACGCGTTTTTGACCAGAACAAAAAAAAAAGAGAAGCCTTTTTTGACCGCCTATTGTCGCTTGAAACATTGTTTTGAATGGTTGGTAATAAACTTCACTTTCCTCTCGGTAGATTTCGTTTGCTGGCTTTGACTCAGACGATTCCCTTAGACCATCTTGATTTTCGCACAAAAACAAGATTTCCGCTCCCATGTGCGCCATATTGAACATGGTGACAATGCTGCCTTAATTCACAAGGAACTCTTCTTTTTTTTTCTGCTTTCAGCTGATATAACGAAAAGCTCGCGTTACGATTTCTGACTGCCTGACGCATCTTATATTTTGGTCCTCGCCTGGTTAACAAACACTGACTACCAAATGGAGCGGATTCTCCAAGCTGTTCAAGAAATGTAAAACACCTATTTTCATTTATGTACGTAGTGATCATCGTAATTAGGATAAATTAGCTATCATTTCATTAGGCTTTTACCTGCTTTTCCGTCTTTTTAAGTTCTGCATTAAGCGACAAAAATGGCCTCTTTACTATTTGGCGATTTGTGTTCAACTAGGCGGGGAAAGGAATTGTTTTTCTGTTTTATGCAAGCAAAGGAAATTTTATTCTCTAGTTATAGTGACCAACATATCTGCAAGGCATTATCTTATTTAAGTGTGAACCTCGTTCCGTGTACGGATCCAGAACTCGATTATTTTCAGCAAAAACTTTCATCCCGTTTCTACCCCTTCGGTTTAATTAGTGAACACAATTTGCTCTACCTGAGGACTTTTCTGCAAGCGAATCGTGGCATTCAATTATCCATCCCAAACATTGACAACCAGGCTTATTTACAGGCTCTTTCGACGCAATCTAGTTTGATGTCGCCGCATTATCTTCCATTACAAAGCAATCAGCTCGGCCAAGTCAAACCAACTTTTCAGCTTTCGGAACCAGGTCGCTCTGATGAAAATTTCAGCGACACTGACCCAAAATTCATTTGCAAGCATCCGAGAATTCACGTTCCAACATCAGTCGGTGTTGTACAGCCTAGTGTAAGACGTCGGACTAGCGACAATCCTGTTAGTCCAACTGAAAGTCGATCTGAATCACCTCTATTTTCACTACTACACGAGCCTGAGACAAGTGTCGCTACAACCACTCTAGGTGATCCGACCAATCAATTAGTTTCGCGAACTCTGGCCAAAAGCAATGTCAACCAACTCTCCGCGCAAGATATTCCTAACGATTCCTCCGTTCAGCAAAACAGCCTCGAAAGTCACCTTACCCCTTTGAACCAACCTGTTGATCCTGATCCGCTATTACCCGAATCCATTGATGGTACCAGCAGCATTGAGATCGATTCTTCTAAAGAAAGTACGAAAAGCAGTAGAACTGTGACTCTTTCCGAATCGGAGATGTCACCCCAGTTGCGTTTAGATTTGGAGGAAATACGAAAATTTTATTCTCTTCCAATTAACCTCAATCGTGACGGAGGTGTTCTGCAAGATGTCTCGATAGGGAAAATGTTGGAAAGGATAAAAGGGTTCTTGTGGTTTTTAAAGAAGGTAAAAGGCGTCGAGCCTGCTTTGACTTATTGTATCAATCCGGAAGTCTTACAACAGTTTGTCGAATTTATGATGAAAAATCGTGGTATCAAAGCCATTACTTGTAGCCGGTATGTGACGTCCTTAATAAGTGCCTGCAAAGTGCCACTCGCGTGCACACAAGATGAACAAAAAGAAGAGTCTCTTGAAAAAATTAGGGCCATTCAGAGGCAACTTGAGCGATTGTCCAGACAGGAAAAAATTGATTCCGACAGTCTTAATCCTCAGACAGACAAAGTAGTTTACTCTGAATTGCTAGAATTATGCAGAGAATTCAAATGGGAGGTTTCGGAAAAAACAGGTGCTGATCGTGCACGAAGTTGTATGAATTTGTGCTTGCTTCTCATGTACTGTGCGGTTAACCCGGGCCGAGTCAAAGAATACATCTCACTGAGAATTTATAAAGATCAAAGCGGCGACCAATTGAAAGATCAAAATTTTATCTGGTTCAAGGAGGACGGTGGCATAGTATTGTTGGAAAATAATTACAAGACCAGAAATACTTACGGCCTAAACACCACTGACGTGAGCTCAGTCACATACTTGAATTACTATCTGCAACTATACAAGTCTAAGATGAGATCACTTTTGCTACACGGCAATGACCACGACTTTTTTTTCGTTGCTCCGAGGGGAAATCGTTTCTCGCATGCCTCTTACAACTATTATATATCCGGACTATTCGAAAAGTACTTATCTCGGAGATTGACAACGGTTGACCTTCGAAAAATTGTTGTTAATTACTTTTTGTCGCTTCCAGAAAGTGGCGATTATTCCTTAAGGGAATCGTTTGCGACTCTCATGAAACATTCTATCAGAGCGCAACAAAAATATTACGATGAACGTCCGTTAACCCAAAAAAAAGATAGAGCGCTCGATTTGTTAACCTCTGTGGCTAGACGAAGTCTAGACGAAGATGAACCTGAGATTGTAAGTGATGAAGACCAGGAAGGATATCTCGACTGCTTACCGGTCCCGGGAGATTTTGTGGCCTTGGTCGCAGCCAATTCTACCGAAAAGGTTCCGGAAGTTTTTGTGGCTAAGGTACTGAGACTTTCCGAGGACAAAAAAACTGCTTATCTTGCCGATTTTGCGGAAGAAGAGCCAGGAAGATTTAAATCGAAAGCGGGAAAAAGTTATAAAGAAAATACAAATTCTCTAATTTTCCCAATTGACATCGTCTTTTCGCATTCGGACGGTCTATATGAATTGAGAACGCCAAAAATTGACCTTCATCTTGTGACAGTTCAAAAGAAAAGTTAA
>FUN_000002
ATGGCTTTCGTTCGCTTGCCGAGGCCATTACTGGACGAATTCATAACGCTGTGGGACCAAGTTCAATCCATTTCGCCGACGCTTCCCGAACAGGCTAGGCATCTTATTCATCGTGTAGATGACAAGGTCGCCGAAATTAGGAGCCAAGCAAGCGACACGCCTACGACAGGAGGTCACGGTGCAGAATCCCAAAGTACAGGTGGGTGAACTAAGCAATGTTAGGTTGACTTATTAAGCAGAACATATGAGGAAATTCATCACTATTATGATCACATCCCTAGAATTTCATACTTCTTTAATGACTTCTAATATTCAGCATTCAATTGTCTTCAGTCATTAATTATAATCTTCTCTCCTTTGAAATGTCTTAATCATTGCACATGCTTTTAATGTTTTTAGCAGCTGACCAGCCACCACAGATCCCTTGCTTGGCCATGCAAGAGGAAACAGCCTCCTCAAGTGGTAAGAGCAGCTGTCTTCCCTGATAAACTATGAGCACTCAGTTCGGTTAACTGTCAACTAAACTCTGACAATAGGCATAGTATACAATAGTTTACTGCCATAGTATGCAATGGGGGAATGTACTGAGAGGAGAGCTGTGAACATCTAAAACATATATCTATATACACCACTTGGAACCAACAACTGACATAAATGTGtaaaaaaaaaatcataataatactaataaataattatGTTGGCATGGAAACTACTGACATGCAGGTGTTTCCATTCAAAAAAATGTCTCAAGATCCAGTCAAGTAAATCAATCTATTGATGAAGAAATTGATATGGTTTAAGAGTTTTCCTGTATTGTACTGATGGGAATGAGACATAACAGCTATCCATCCTCTTCATATAATTTTTTTCTCAAAAATAACCATAGATTCCAACCTTTTGGAAGGTTACATCCTCATAAAGGGATTACTGTGAGGACAGCAAGCATTTGTAAAAAAAATATTTATATATTTACCTTTCTTGGAGCCAAATCAGCTATCATAAATCTCCACTCTTTATGTTGGCAAGTAAACAACTGACATTAAGGTGTTACTTTCAGTACATGTCTCAAGATCCAGTCAAGAGGAGCATGATGATGAAATTGGTATGGTTTAAGAGTTTTCCTGCATAATTTGTACTGATTGAATGTGTTAAGGATGACAGCCATTGTATCATTAAATTTTGTCTTTAAATGTTATTTTCTTCTGAAAAGGTCTTTTATGTAACAGCAAGAAATGCTGTAATGAAAGAAGTGCTGAATCTTAGCAAACATCACTGATATCATAACTTTTAATCCCTCCATCAGATGACCAGCCAGCAACTCCACTGGTTAGGAAGCGCAAGAGGAGAGTTGCTCAGAAAATAACAAGCTCTGCTCAGGTAATTGAAAGAAATTTTACTTCAATAATACTATTATTGCTGGTGCGACCCTGTACACACAGATTTATATTGAAGAGAACAAATAACCTGGGCCTCCATCCCCCAATTAACACAATATCTACCCCACCACTTCCCCTGTAGAACCACAGTTCACTGGAAAATAGATCAATAAAAGAATCAAGATTGAATGATTTGCCATGATTTATTCTCCAGTAAACAACAAAAACAGCAACAAAAGCAAAATAATATTATTATTCTGACTCCTGGATTTCATTGAGACTTCAAGTAACTGGCCATCATTTACTTATCTATTTTTTTAAGTGTCTTAGTGTAGTCCACCGTAATAATAATGTTTGCTGCAACTGCAAGACAAAATTTCTCTTCCAAGAAATATATCTTCAAAATGCCTTACTGAAGAAACTAAGATACTTCTTTCCCATAGTCAATCAATTAAGCAAATAAAAAAGTAACATTTTGTTCATGGTAAAATTGCTGTCATTTAACTCCTGACTATGCTAGTGAAGTTTATTTGTATTGTCCGCATTTCCTGAGGATAAGAGCCTTTCATGCAAATTTACTACATAGTTCTGAAGACGTTAAGCAGACAGAAATGAGCCAAAATTAATTTTCTTTCCTTTTAAACTTATAGGGCATCCAGTTGCCAGCACGGCCACCCGACATCGTGTCAGAGCCCACCCCTGACATAAACTTTGAAAGCATGGAGATCAAGTATCTCAAGCAAGCCTTAGCCTCTGTGGTACAACCATCAACCAAGGCAGAGGTTTACTTGAAATATCTTGCAAAATGTGACTTCAATACTACATTGGAGCCACACTCCATTATCAAGTTTAATCAGGATGATGTTCGGAGAATGGTTGGAATTGGGCAAAATCCCAATGGTGAAGAAGAAGTAAGACAATTTTTAAATAAAAAGCAACACAAATGCAGTGCAAAGTACCTTTGAAGGAATTTCAGTCGAGAATAAGTTCAATCTTTCAAAATTAGCGAGGGGCTCTGATGGTGCTAATCACTGCTTTTCATGGGAATGACCTCTGCATGATAGTTTGCACTGCATTAGCCAATAAAAAAAATTAATACAAACTCATTGCCTGAGTTGTAAAATTATTTTCGAAATAACTTATACCCTATTCACACCAACTAAACATGTCTAATTAAACTCGCTTTAACAAAGAAAATCAGTCCAAAAAAAGTATGGGACTGGTTTTTACAAAAGACCCTGATAAGTTTTAATTTATATGCATTGATTGATATGTACTAAATTTGAAGTTGACAAAACATCATTGCACACGATTTGATGTGAAGACAACTTAAAACCATGTTTAACTAAACAGCTTTTAAACATGTTAGAGCTTTGGTGTGAATAAAGCTTTAGTCACAGACAGTCATGAGTTCATGAAGTATATTCCACAAAATAAAATTGTCTCTTTCTTTTCTCACTGTTTCCTATCCATTGATTGTTAGGTTGTTGGAAAATTACACCAAGCCCTCCAACAGAGATTGCAGCTGGCTGTCCAACAGGCATTTTCAATAGGAGAATTCATGTCGACGTGCATCAAGGAGCATGGATCAACTTTGCCAGATGAGGACCATCAGCGACGCCGTGGAAGACCTCGAGTTTCATCATTATTAGACACGCTGGATTCCATCGAGTCACTTGGAGCTTCATCATCGTGTCTACAGGACCAAGTTTTGATTTTTCAAGCTATTTCAAATTTTCCATTATTGAAATATGTTAATGAGCCTATGACTCACTTTGTGTGTAATGACGTCAGGTTAGCGATTAGGCATCTACCAGCAGCATTGCATCACAAGTGTGATGTACCAGAAAACATCCAAGATGCCCATATCAACTTGAGACCGTTTAGGTCTTTGGTGTCAGATCAAGAAAGTCCACCGCTTTCGGGTGACATGAGTTACTCCAATGTAACTCCATGTGATGGTGATGGTCTCGTTGTTGGTATTAATCTTGACCACCGAGAACCACTGGAAAATGCTGAATATACACGCTTTTACGGTATCGATGCCCCTGAGTTGTCATCTGTTCATTTTATTAAAACTAATGACTTCCAGCATGTTTTCTGTAAGCAGGTTGGGCACATAAGCCTCTGTGCGGTACATTTGTTTTTACAGATGTTCTTGTTGAGTGGATCTGCAAAGCTATGCGAGGAACTCCCCCGGGAAGCGGCTCCACAGCCAAGGGACATTTACAACAGAGCACTCAAGGAGTATTGGTTCAAGTTCATCACACCTCCGTCTCAACATCTGGAGAAAGTATTCCTGCAGTCTCTTGAAGAGCTAGTCCCTCCAACATCTGAATCCCGAAAGAGGCTCATGTCGCCTTTTCCAGCAACCATGGCAACTGCTGCCAATCCATTCTTGTTGTCACTAAATGCTCTTCTTGTTGTTTCAGGTTTTTGTCACGTCTTCACGAAGTATTGCCAAGATGGATTTCTTTTAGGGCTGCAGGCAATTGCACGGGACAACAAGCTAGGGCCAATTTGGTGTGGTGCTTCTCGGAAGTTCATTTTTGGGTGTACTTCTGGAAACAACACTGATTTTTTTTTGAAGCACTTTACACCAGAAACAACATCTCACTTGGCCAGGGCTGGTTTTCCCTTCAAACACTCAAATGCTTTTCTCCCGTGGCACGAGAGACAAATGCTGAAACAGCTGTGCTCACAGGAGACTACAAGAACCGCAGCAAGAAATCACTTGGCACAGCATCTCCCAGGAATGGAACCCCAGTTTGGCATGTATATTGACATCCAGAGGTTGGTGATTATTGTTGAATAATAACTACTACCCTATTCACACCAACTAAACATGTTCAATTAAATTCGGTTTAACAAAAGAAAATCAATCCAAAAAAGCAAAAGACCCTAGTAAGTTTTAAATTACATGCATTGATATGTACTAAATTTGAAATTGACAAAAATCATTGCACACGATTTGATTTGAAGACAACTTGAAACCATGTTTAACTACAAACAGCTTTTAAACATGTTTAAGTTTTGGTGTGAATAGGGCTTATAATTCTATTATTTTTTCATTGTTACCTAATTGTGACCTCATGATAACATCATCACATTTTTGTCAATAAAATTTGTAGATCTAACCAAGGAGAAGGATATCAAACAGTGAGGACTGGTGAGGCTTACCTTAAAGTTATGAATAGCCAGGTAGTTGGGATAGGAGGTAATTCTACAGGACTGGGTTTGTTCACCCTTAAAAAGATTCCTAGAGGCACTCTTGTATGTGCCTATGCTCCAACTGCAACAATTTGGGAAGGTAAACTAAATGGTGATTATGTTTTAGAAACTTCTTTTAATAACAAGGTAATTTCTGTCAATGGGAAGGAAAATCTTTTTGAATTGGGGTTGGGAATTTATTGTAATGATGGAAGCTTTCCATTTTCCTTAGCTAGGGCTAGGTTTTCAAATGTTATTTCACATAGAGTAAATTGTGAATACTGTAAATGTGGGGATGGCATATGGCTAAAGACCGTCAGAGATGTTTCCGCAGGGGAAGAGCTGCTGATGTGCTATTCTCAAGATGGAAGCTACTGGGCTACTATCTTCAGTAGGGAACAACTTAACCAGATCACAGCTGCCTTAAACTCATGTGGCCCATCACTTCAGGATGCCGAGCGCTGCATTAGACTACTTCAAGTCTAA
>FUN_000003
ATGTCCAACTTCAGTATAAAGGTTGGTTGTATAGAAGGAAAGTGTTCGCTTTCATTTCATATTTATATTTGAAAATATTTTTATTGTCATAGTTTTTTTTCATTGCTCATTCCTGGTTTAGATATATCAGTATGCTCCGGCGGAAAAGAGAAGGTTTTCTGCCTGCGTGCGCATTTTGCGTCGGTAACGAAGAAATCAATTGACTGTGCTGTCCGGCACATCTTGGGTGTGAGAAGAAGATCTTGAACCTCTCTCTCCATATTTGCTTTGCTTCTGTAGTCTTTTTATTGGTAATTCCTGATCAGTTGGCCGTTTTGTATAGGTAAAAACGGTTGCTATTGAAAGAATATTTTAGGAGCATTTGTTTTTACAGATGTTCTTGTTGAGTGGATCTGCAAAGTTATGTTTGTTTCACGAAAAGCGAGAAGAATCCTAATTCTTTGAGGTTGATTTTTGGTTTTTGACTTGTGGGACAGCTGCTATCGCAACTGTGGTAGTGCGCGCATTTTGCGCAAATCAGTTGGCCGTACTTTCGTATGATGAAAACCTGTTCACATTCGTTCATAAACATGTCTCCAATGGGACCCGGGCTTATCTCATTGATCCAGCTTTCATTTCAGAGATTCGGAGTGTCTTAGTAATGGTCGCCATTTTTACCTGGAAAATTCGGCCTACTATGTTACGGAAGAACAGAGCAGTGTCCAATGCAATTTCAAAACAGGCGATTGATACCTTCTTGACGGTATTCTAGGCTCGGGCCTTGATGTAGTCATACAGCTTGCTAAGAAAACTTTAAATTGAGCCGATTGGAGCGTCCTCTTAGCGTGCAAATTCtttggtttttgccgttctgtttttgctttttgttttaatttttAAAAATTTCTCCGTGCTGAGTAATAGAGGCGTCATGTTCCGATGGCAATGGATGAATTCTCGTTGGCCGTTACTGCTTGCAATGCAGTTCTCCTTGCTCCTCTTGTCTTCATTGTTTGTTGGGTTGTGGGAGAACGAATGACACCCCGCTTTGTTACATGCTTAACTGTATTAATTTTTCATCCCACAGTCAGAAGATGAGCCGGCAAGCCCTTTGGTCAGAACTGATAAGAATGAAGAGTTACCCACAATACCACAAGCTTTCAGGAAGCTAAAGGAGTGTTACTGCAACTGTTCTTGGGGAGATTCTTTACATGCAGATCCTCAGTTAACTGGAGAGTAA
>FUN_000004
ATGCTCATTTTCAATCGAGGTATTGTCTATGTTGTCATGAACAGTGTGTGTTTAATAGGAGTTTGAAAAAATAAGAGCCCCATTGAACACTCGTACATATCTATTGTTGGCTTAAAGAGAACTCTTTGGCGTTTATATAAACGAATCGTAGATCATGAGAAGATGGCGCTAATCCCTCTATTACGTTCGCAGACCCTCATGATAAATGTGTCAGCCAGACAAAATCGAAATTTTTGGCAACGGAATGCCAGATAGCcagggctcagttgttaaaatccggattaagctaatcatggattagaggaaattttagttgttatttatttaccgttaaagggggctttttcataggattatggcttaaggaaaagaaatttgtaatttataaccttattgggtcccaatgttgtggcaaaacctccttttagccgtaaataaatagcaattaaaataacctaggattagcttaatcgagcttcgaacaactgggccctggtctcttgagagAACACGGCGATCCTGGGTACGAACGATACAATGCAAATGACTTATTTTTCGACCAATACGAGATGACTCCACGTGACTTGAATTCTTTCCGTTGAGGCGGCGTGGCTTCAACGGTCTTTCTTCAATCGTATTTACACATGACTTTGATGTACATCATGCAACAAGAAAGTGACAATCAAGGCAAACCGACTAACAGCAAATTGCAGTTATTGCAAACTCATGCAAGAGGTGTCCATGTGTAACACTCAGTGGATTGCCAAAAGTTGTCTTTCAGAACACTCACGACAATGAGAAGATTTAGATTTAACCATTTCATGCACACATTATGAATCTTGCAGACCTTACCTTCGACACATCTTCAAATCAGAGATAAACATTTACGAACATTTCAACATGGTATTAAAGACCCTTCTCTCCCCTGCCAAGGGAATTCAATTGTTACGATttgttgttgttgtcgttgttTAAATATTTTATAGTTACTTTTCGAGTTAAACATTGACTTCAAAAACTCTTAAAAACCTGGAATTTCAAGTGTTTAGATTTTGAAAAAGAGCCTGATATATCACAAAAAATTTTCCAGTTATAATTAATTAGTAACATTTCCATATCGTCCAAAACTGAGACTCAAGTCAACTAACAACTAACAACTAATTCTTTTCTCTTGTTAACTGATGAAATATAATTATGTAACACATGTTCTGTTGACCTTTGGATAATAAAATGTGTTGTTTGTTATCTTCGCCATAATCAATATGTTAACCGTTATCATCAAAAATAGAAGCCTGAAATTTTATGTCAAGCCCGGAAATAACTCATTTCGCACTCTTTACTCTAGGATCGCCCTTAATTGAAAGCACGTACGTGCGGCTTGTGGACATATTTCCGGTTCTGGCAGTTCATTGCCCTCTGTGGGGCATTAACCTAATCAACAACAAATGCAAAGGTGACATGCTCTGATCGGAAGGAACTCTGACACGCTTTAAAGCTACTCTGCTGAATATGCTCCGACCTTTTTAAAATTTATTTAAGTCAGAGAGTGTCCTGACTCAAAAGCTTTTGAAAGGAAACGTGAAAGGTTGTCTACTGTTAATCGCGCAACGGAGATGTCTCGTAAAGTTTCAATGGCGGTGATTTAATTAAAGGCCAGAAATAAACATGTAGGTACCTCGTAAGGTGCTAGCGGTGGTGTTTAATGTCTCTCTAGTTGTGAAACTGGTTAAAAAAGGCCATAATAATAGGTTTCCTTTCTACTCCTTCGCGGAACTGAGCGGGTTTCTCTGTGTTAGTTTCTACTTCTCCCTTTTTGGTCTGTTTTCGAATAATGTGGCTGTGACAGGGAGGATGCAAAACCACCATCAGTGCTATGGTATTCCTTGACTTATATCAACGTTGACCTTTTACCTGTTAGTCATTGCGCACGAGCTACAATAAATTCATTATGATTTTTGTCTGCTTTTCCTGTTTGCCGCAGGGGAGCTTCTGTTTTTACCAATCAGGAACGTTCTCTGCATGATGCCACAAGGGTACAAAAACGCCCTTCCAGGCTATAAAGACCTCTATCTTAGCCAAGCAATCACCGAGGAGGTTCACAATGTAAGTTTGTCCTTTTAGCTTGTGCCTGTTtttcctagtttgtttgcttgtttctttctttctttctttctttttagttctattttgtttcattTGTGATTCTTCAAGATATTTGGTGGTTACTTGTTCAGTCTAGCGTAGCCAAACTATTTGAACACTACAAGATAATTCGCAAGAAACAGGTTGAAGAAAATTCCATTTCAAGCGAAATCTTATTTCATTTTTTTACATTCTCTGTAGCTAATATACTGGCAAGTAATTTTATCGCCAACTGAAGGCAAGCACCAAGGACGTTTACATGGTAAGTTTGTCCTTTTGTTACACAAATTGGGTAGTCTGTTTCCTGGTGTTTTTTTTTTTTTTAATGTTTTAATTCACGATTATTCAGGATATCTAGGGATGAATTATCTGTTTATTACTCCAACATAATTAGTAAGAAACCAATTGGGAAGAATTCCAATAGAAGCAAAATCTTAATCTGTCTTTAAACTACTATGTAGTTAATACTTTAGCGAGTGATCTCTTCTCGGTGGCTATTACCATTTGTTGTGAATGCTATGAACGAAGAGCACCTTATTTTACTACCCGACATTATTGTAGCCCTGAGTTCCCTTTAGATCTGACGCTGCCATTTCTTACTTTAAGGCCTCTTCAATTTCTTGGTATTCGTAGATGTTTTCCACTGGCATTGACTGTGGTGTAAGTTCTTTCAAGCACGGGCCGTCGTTGTCTCTTCTAGCACTAGACAAAAAGGTGAGTGAGTTAAAACATGTTGGCTGTTAGTTCTTTCTACTATTTTCAGTTTTGCACTTGAATTCGTTGGAAGCCTCTGTGACAAGGCTTGATTTAATACTCGCTTTTTGGCATTTCAATTTCGCTTTCACCAAGAGAAACTCGATATTTCGTTGACTTCGCAATAAGTTAGTAGTTTTTCTTTCATATTGAGAGCTTCGCATTAAGGGTTCACATCGATGCTCTTGACTAGGCAAAGAAACGAGTCATTGTGCGAATTGCAGCATCTGTTTCCTACAATCACATATCTACACATATTTCTGGTGGTTTTAATTATAGTATCAATCAATCATCTTGGTTGAATATTGGTTAGGCAAAAGAGCATCCCAAAAAAACGTTTCGATCGAGCGTGTGACCAAACGTGTGGTATGACGGGATTCCTGGTATTGCAATGACCACTGATTTAACACATGAAATGGATACCTTAGAAATATTTCCTGTGATAGTGGTTTTTAAGACATTGTGCTAAATGCCACCCTGGATACTTTATCAACAGCAATTTTGGCCTCAGTAGGCACGTTTAGTAACCTCGAATTAACGCCACTTAATTTTTTTGGCAGCTCTGTGTTATCATATTCAAGTTATTACTCCAAAGCCACCTTCCCCTGTTTTGTCTGCGATCGTTGAACACAACGGTGAGTGAGAACTGAATATCAATATTACTATTAATATATCTTGCTTATAGCAGTAATTGAATTTGCTGCGGATGTACAACAATACTTATATTGTTGTTTTGCACCTCAATTAAGCTCAATTTAACGTCACTTAACTTTCTTGGCAGCTCTGTGTCATCGCATTGATCGAGTCATTATTCCAAGTACGTGGCCTTCCCTTCTGCGCACGATCGCTGAACAAAACGGTGAGTGAGAAAACTGAAGCGTTACATGGAAACTAACTGTAGTCAAGTCGCATCAAGGTTTCACCCTTCTAGAACGACTCAGTTAGCTGTCGTTTTGCTTCAAGTTTTTTTCCGGCAGTACTTAGTAGCATAATTTTGACCATTGACCGCTAATTATTGTGCTCAAAATCTCTCCGCAGATTTCTTTGGTGCGTTTGGCTGGGGCATTTCACCGTAAACACGAGCCCCCTTTGACTCCGTGTCCTCGAACGTCGCAAGAAACAGTGAGTAAAACACTTTGTCTGTATGATTCAACTCCATCAGCTGCATTTGCATGTCAGAAAAGCGTTAATTATCGTTTCATTTCCCTTTTATCTACTTTCAATAACTTGTTTCTTTAGAAAAGAAATTTTATATTCGATAGGCTATCTCACTATTTAGAAAAGAGACAACTTACCTTTGGCAAGCAATTTGATTTTCACTCCTATCATTCAATTGATCATGCCATGTTAAGTATTATTCATAAAATCAATGACCGCTAATTATTATGCTCAAAATCTCCCCGCAGATTTCTTTAGTGCATTTGGACAGGGCGCCGTTTCACCGTAAACACGAGCCCCCTTTGATTCCGTGTCCTCGAACGTCGCAAGAAACGGTGAGTAAAACACTTTGTCTATATGATTCAACTCCATCAGTTGCATTAAATATGTCAGAAAAGCGTTAATTATCGTTTCATTTCCCTTTTATCTTTTTTCAATAACTTGTTACTTTAGAAAAGAAATTTGCATTCGATAGGCTGTCTGACTATTTAGAAAAGAGACAACTTACCTTTGGCAAGCAATTTGATTTTCACTCCTATCATTCAATTGATCATGCCATGTTAAGTATTATTCATAAAATCAATGACCGCTAATTATTATGCTCAAATTCTTCCCGCAGATTTCTTTAGTGCATTTGGACAGGGCGCCGTTTCACCGTAAACACGAGCCCCCTTTGACTCCGTGTCCTCGAACGTCGCAAGAAACGGTGAGTAAAACACTTTGTCTATATGATTTAACTCCATCAGCTGCATTTATATGTTAGAAAAGCTTTAATTATCGTTTCATTTTCCCTTTTATCTATTTTCAATAAGTTGTTGCTGTAGAAAAAAAAAAACTTGTATTCAATAGGCTATCTGACTATTTAGAAAAGAGTCAAGTTACCTTTAGTAAGCAATTTGATTTTTGCACCCATGCAACATTCATCTGATCATGCCAAGTTAAGTATTGGTGATCAACTGCAAAAGGCAATTGCTGGAGATTTTTTAGGCTTGAGCAAAGCTTATTTACCTAGAAAGAAGTCACGTTCTTCCTCACACTGAAAACATGCCAACTTTCTTTCTTTTTCTGACAAGAACGAACGCGTTTTTGACCAGAACAAAAAAAAAAGAGAAGCCTTTTTTGACCGCCTATTGTCGCTTGAAACATTGTTTTGAATGGTTGGTAATAAACTTCACTTTCCTCTCGGTAGATTTCGTTTGCTGGCTTTGACTCAGACGATTCCCTTAGACCATCTTGATTTTCGCACAAAAACAAGATTTCCGCTCCCATGTGCGCCATATTGAACATGGTGACAATGCTGCCTTAATTCACAAGGAACTCTTCTTTTTTTTTCTGCTTTCAGCTGATATAACGAAAAGCTCGCGTTACGATTTCTGACTGCCTGACGCATCTTATATTTTGGTCCTCGCCTGGTTAACAAACACTGACTACCAAATGGAGCGGATTCTCCAAGCTGTTCAAGAAATGTAAAACACCTATTTTCATTTATGTACGTAGTGATCATCGTAATTAGGATAAATTAGCTATCATTTCATTAGGCTTTTACCTGCTTTTCCGTCTTTTTAAGTTCTGCATTAAGCGACAAAAATGGCCTCTTTACTATTTGGCGATTTGTGTTCAACTAGGCGGGGAAAGGAATTGTTTTTCTGTTTTATGCAAGCAAAGGAAATTTTATTCTCTAGTTATAGTGACCAACATATCTGCAAGGCATTATCTTATTTAAGTGTGAACCTCGTTCCGTGTACGGATCCAGAACTCGATTATTTTCAGCAAAAACTTTCATCCCGTTTCTACCCCTTCGGTTTAATTAGTGAACACAATTTGCTCTACCTGAGGACTTTTCTGCAAGCGAATCGTGGCATTCAATTATCCATCCCAAACATTGACAACCAGGCTTATTTACAGGCTCTTTCGACGCAATCTAGTTTGATGTCGCCGCATTATCTTCCATTACAAAGCAATCAGCTCGGCCAAGTCAAACCAACTTTTCAGCTTTCGGAACCAGGTCGCTCTGATGAAAATTTCAGCGACACTGACCCAAAATTCATTTGCAAGCATCCGAGAATTCACGTTCCAACATCAGTCGGTGTTGTACAGCCTAGTGTAAGACGTCGGACTAGCGACAATCCTGTTAGTCCAACTGAAAGTCGATCTGAATCACCTCTATTTTCACTACTACACGAGCCTGAGACAAGTGTCGCTACAACCACTCTAGGTGATCCGACCAATCAATTAGTTTCGCGAACTCTGGCCAAAAGCAATGTCAACCAACTCTCCGCGCAAGATATTCCTAACGATTCCTCCGTTCAGCAAAACAGCCTCGAAAGTCACCTTACCCCTTTGAACCAACCTGTTGATCCTGATCCGCTATTACCCGAATCCATTGATGGTACCAGCAGCATTGAGATCGATTCTTCTAAAGAAAGTACGAAAAGCAGTAGAACTGTGACTCTTTCCGAATCGGAGATGTCACCCCAGTTGCGTTTAGATTTGGAGGAAATACGAAAATTTTATTCTCTTCCAATTAACCTCAATCGTGACGGAGGTGTTCTGCAAGATGTCTCGATAGGGAAAATGTTGGAAAGGATAAAAGGGTTCTTGTGGTTTTTAAAGAAGGTAAAAGGCGTCGAGCCTGCTTTGACTTATTGTATCAATCCGGAAGTCTTACAACAGTTTGTCGAATTTATGATGAAAAATCGTGGTATCAAAGCCATTACTTGTAGCCGGTATGTGACGTCCTTAATAAGTGCCTGCAAAGTGCCACTCGCGTGCACACAAGATGAACAAAAAGAAGAGTCTCTTGAAAAAATTAGGGCCATTCAGAGGCAACTTGAGCGATTGTCCAGACAGGAAAAAATTGATTCCGACAGTCTTAATCCTCAGACAGACAAAGTAGTTTACTCTGAATTGCTAGAATTATGCAGAGAATTCAAATGGGAGGTTTCGGAAAAAACAGGTGCTGATCGTGCACGAAGTTGTATGAATTTGTGCTTGCTTCTCATGTACTGTGCGGTTAACCCGGGCCGAGTCAAAGAATACATCTCACTGAGAATTTATAAAGATCAAAGCGGCGACCAATTGAAAGATCAAAATTTTATCTGGTTCAAGGAGGACGGTGGCATAGTATTGTTGGAAAATAATTACAAGACCAGAAATACTTACGGCCTAAACACCACTGACGTGAGCTCAGTCACATACTTGAATTACTATCTGCAACTATACAAGTCTAAGATGAGATCACTTTTGCTACACGGCAATGACCACGACTTTTTTTTCGTTGCTCCGAGGGGAAATCGTTTCTCGCATGCCTCTTACAACTATTATATATCCGGACTATTCGAAAAGTACTTATCTCGGAGATTGACAACGGTTGACCTTCGAAAAATTGTTGTTAATTACTTTTTGTCGCTTCCAGAAAGTGGCGATTATTCCTTAAGGGAATCGTTTGCGACTCTCATGAAACATTCTATCAGAGCGCAACAAAAATATTACGATGAACGTCCGTTAACCCAAAAAAAAGATAGAGCGCTCGATTTGTTAACCTCTGTGGCTAGACGAAGTCTAGACGAAGATGAACCTGAGATTGTAAGTGATGAAGACCAGGAAGGATATCTCGACTGCTTACCGGTCCCGGGAGATTTTGTGGCCTTGGTCGCAGCCAATTCTACCGAAAAGGTTCCGGAAGTTTTTGTGGCTAAGGTACTGAGACTTTCCGAGGACAAAAAAACTGCTTATCTTGCCGATTTTGCGGAAGAAGAGCCAGGAAGATTTAAATCGAAAGCGGGAAAAAGTTATAAAGAAAATACAAATTCTCTAATTTTCCCAATTGACATCGTCTTTTCGCATTCGGACGGTCTATATGAATTGAGAACGCCAAAAATTGACCTTCATCTTGTGACAGTTCAAAAGAAAAGTTAA
>FUN_000005
ATGGCTTTCGTTCGCTTGCCGAGGCCATTACTGGACGAATTCATAACGCTGTGGGACCAAGTTCAATCCATTTCGCCGACGCTTCCCGAACAGGCTAGGCATCTTATTCATCGTGTAGATGACAAGGTCGCCGAAATTAGGAGCCAAGCAAGCGACACGCCTACGACAGGAGGTCACGGTGCAGAATCCCAAAGTACAGGTGGGTGAACTAAGCAATGTTAGGTTGACTTATTAAGCAGAACATATGAGGAAATTCATCACTATTATGATCACATCCCTAGAATTTCATACTTCTTTAATGACTTCTAATATTCAGCATTCAATTGTCTTCAGTCATTAATTATAATCTTCTCTCCTTTGAAATGTCTTAATCATTGCACATGCTTTTAATGTTTTTAGCAGCTGACCAGCCACCACAGATCCCTTGCTTGGCCATGCAAGAGGAAACAGCCTCCTCAAGTGGTAAGAGCAGCTGTCTTCCCTGATAAACTATGAGCACTCAGTTCGGTTAACTGTCAACTAAACTCTGACAATAGGCATAGTATACAATAGTTTACTGCCATAGTATGCAATGGGGGAATGTACTGAGAGGAGAGCTGTGAACATCTAAAACATATATCTATATACACCACTTGGAACCAACAACTGACATAAATGTGtaaaaaaaaaatcataataatactaataaataattatGTTGGCATGGAAACTACTGACATGCAGGTGTTTCCATTCAAAAAAATGTCTCAAGATCCAGTCAAGTAAATCAATCTATTGATGAAGAAATTGATATGGTTTAAGAGTTTTCCTGTATTGTACTGATGGGAATGAGACATAACAGCTATCCATCCTCTTCATATAATTTTTTTCTCAAAAATAACCATAGATTCCAACCTTTTGGAAGGTTACATCCTCATAAAGGGATTACTGTGAGGACAGCAAGCATTTGTAAAAAAAATATTTATATATTTACCTTTCTTGGAGCCAAATCAGCTATCATAAATCTCCACTCTTTATGTTGGCAAGTAAACAACTGACATTAAGGTGTTACTTTCAGTACATGTCTCAAGATCCAGTCAAGAGGAGCATGATGATGAAATTGGTATGGTTTAAGAGTTTTCCTGCATAATTTGTACTGATTGAATGTGTTAAGGATGACAGCCATTGTATCATTAAATTTTGTCTTTAAATGTTATTTTCTTCTGAAAAGGTCTTTTATGTAACAGCAAGAAATGCTGTAATGAAAGAAGTGCTGAATCTTAGCAAACATCACTGATATCATAACTTTTAATCCCTCCATCAGATGACCAGCCAGCAACTCCACTGGTTAGGAAGCGCAAGAGGAGAGTTGCTCAGAAAATAACAAGCTCTGCTCAGGTAATTGAAAGAAATTTTACTTCAATAATACTATTATTGCTGGTGCGACCCTGTACACACAGATTTATATTGAAGAGAACAAATAACCTGGGCCTCCATCCCCCAATTAACACAATATCTACCCCACCACTTCCCCTGTAGAACCACAGTTCACTGGAAAATAGATCAATAAATGAATCAAGATTGAATGATTTGCCATGATTTATTCTCCAGTAAACAACAAAAACAGCAACAAAAGCAAAATAATATTATTATTCTGACTCCTGGATTTCATTGAGACTTCAAGTAACTGGCCATCATTTACTTATCTATTTTTTTAAGTGTCTTAGTGTAGTCCACCGTAATAATAATGTTTGCTGCAACTGCAAGACAAAATTTCTCTTCCAAGAAATATATCTTCAAAATGCCTTACTGAAGAAACTAAGATACTTCTTTCCCATAGTCAATCAATTAAGCAAATAAAAAAGTAACATTTTGTTCATGGTAAAATTGCTGTCATTTAACTCCTGACTATGCTAGTGAAGTTTATTTGTATTGTCCGCATTTCCTGAGGATAAGAGCCTTTCATGCAAATTTACTACATAGTTCTGAAGACGTTAAGCAGACAGAAATGAGCCAAAATTAATTTTCTTTCCTTTTAAACTTATAGGGCATCCAGTTGCCAGCACGGCCACCCGACATCGTGTCAGAGCCCACCCCTGACATAAACTTTGAAAGCATGGAGATCAAGTATCTCAAGCAAGCCTTAGCCTCTGTGGTACAACCATCAACCAAGGCAGAGGTTTACTTGAAATATCTTGCAAAATGTGACTTCAATACTACATTGGAGCCACACTCCATTATCAAGTTTAACCAGGATGATGTTCGGAGAATGGTTGGAATTGGGCAAAATCCCAATGGTGAAGAAGAAGTAAGACAATTTTTAAATAAAAAGCAACACAAATGCAGTGCAAAGTACCTTTGAAGGAATTCCAGTCGAGAATAAGTTCAATCTTTCAAAATTAGCGAGGGGCTCTGATGGTGCTAATCACTGCTTTTCATGGGAATGACCTCTGCATGATAGTTTGCACTGCATTAGCCAATAAAAAAAATTAATACAAACTCATTGCCTGAGTTGTAAAATTATTTTCGAAATAACTTATACCCTATTCACACCAACTAAACATGTCTAATTAAACTCGCTTTAACAAAGAAAATCAGTCCAAAAAAAGTATGGGACTGGTTTTTACAAAAGACCCTGGTAAGTTTTAATTTATATGCATTGATTGATATGTACTAAATTTGAAGTTGACAAAACATCATTGCACACGATTTGATGTGAAGACAACTTAAAACCATGTTTAACTAAACAGCTTTTAAACATGTTAGAGCTTTGGTGTGAATAAAGCTTTAGTCACAGACAGTCATGAGTTCATGAAGTATATTCCACAAAATAAAATTGTCTCTTTCTTTTCTCACTGTTTCCTATCCATTGATTGTTAGGTTGTTGGAAAATTACACCAAGCCCTCCAACAGAGATTGCAGCTGGCTGTCCAACAGGCATTTTCAATAGGAGAATTCATGTCGACGTGCATCAAGGAGCATGGATCAACTTTGCCAGATGAGGACCATCAGCGACGCCGTGGAAGACCTCGAGTTTCATCATTATTAGACACGCTGGATTCCATCGAGTCACTTGGAGCTTCATCATCGTGTCTACAGGACCAAGTTTTGATTTTTCAAGCTATTTCAAATTTTCCATTATTGAAATATGTTAATGAGCCTATGACTCACTTTGTGTGTAATGACGTCAGGTTAGCGATTAGGCATCTACCAGCAGCATTGCATCACAAGTGTGATGTACCAGAAAACATCCAAGATGCCCATATCAACTTGAGACCGTTTAGGTCTTTGGTGTCAGATCAAGAAAGTCCACCGCTTTCGGGTGACATGAGTTACTCCAATGTAACTCCATGTGATGGTGATGGTCTCGTTGTTGGTATTAATCTTGACCACCGAGAACCACTGGAAAATGCTGAATATACACGCTTTTACGGTATCGATGCCCCTGAGTTGTCATCTGTTCATTTTATTAAAACTAATGACTTCCAGCATGTTTTCTGTAAGCAGGTTGGGCACATAAGCCTCTGCGCAGTACATTTGTTTTTACAGATGTTCTTGTTGAGTGGATCTGCAAAGCTATGCGAGGAACTCCCCTGGGAAGCGGCTCCACAGCCAAGGGACATTTACAACAGAGCACTCAAGGAGTATTGGTTCAAGTTCATCACACCTCCGTCTCAACATCTGGAGAAAGTATTCCTGCAGTCTCTTGAAGAGCTAGTCCCTCCAACATCTGAATCCCGAAAGAGGCTCATGTCGCCTTTTCCAGCAACCATGGCAACTGCTGCCAATCCATTCTTGTTGTCACTAAATGCTCTTCTTGTTGTTTCAGGTTTTTGTCACGTCTTCACGAAGTATTGCCAAGATGGATTTCTTTTAGGGCTGCAGGCAATTGCACGGGACAACAAACTAGGGCCAATTTGGTGTGGTGCTTCTCGGAAGTTCATTTTTGGGTGTACTTCTGGAAACAACACTGATTTTTTTTTGAAGCACTTTACACCAGAAACAACATCTCACTTGGCCAGGGCTGGTTTTCCCTTCAAACACTCAAATGCTTTTCTCCCGTGGCACGAGAGACAAATGCTGAAACAGCTGTGCTCACAGGAGACTACAAGAACCGCAGCAAGAAATCACTTGGCACAGCATCTCCCAGGAATGGAACCCCAGTTTGGCATGTATATTGACATCCAGAGGTTGGTGATTATTGTTGAATAATAACTACTACCCTATTCACACCAACTAAACATGTTCAATTAAATTCGGTTTAACAAAAGAAAATCAATCCAAAAAAGCAAAAGACCCTAGTAAGTTTTAAATTACATGCATTGATATGTACTAAATTTGAAATTGACAAAAATCATTGCACACGATTTGATTTGAAGACAACTTGAAACCATGTTTAACTACAAACAGCTTTTAAACATGTTTAAGTTTTGGTGTGAATAGGGCTTATAATTCTATTATTTTTTCATTGTTACCTAATTGTGACCTCATGATAACATCATCACATTTTTGTCAATAAAATTTGTAGATCTAACCAAGGAGAAGGATATCAAACAGTGAGGACTGGTGAGGCTTACCTTAAAGTTATGAATAGCCAGGTAGTTGGGATAGGAGGTAATTCTACAGGACTGGGTTTGTTCACCCTTAAAAAGATTCCTAGAGGCACTCTTGTATGTGCCTATGCTCCAACTGCAACAATTTGGGAAGGTAAACTAAATGGTGATTATGTTTTAGAAACTTCTTTTAATAACAAGGTAATTTCTGTCAATGGGAAGGAAAATCTTTTTGAATTGGGGTTGGGAATTTATTGTAATGATGGAAGCTTTCCATTTTCCTTAGCTAGGGCTAGGTTTTCAAATGTTATTTCACATAGAGTAAATTGTGAATACTGTAAATGTGGGGATGGCATATGGCTAAAGACCGTCAGAGATGTTTCCGCAGGGGAAGAGCTGCTGATGTGCTATTCTCAAGATGGAAGCTACTGGGCTACTATCTTCAGTAGGGAACAACTTAACCAGATCACAGCTGCCTTAAACTCATGTGGCCCATCACTTCAGGATGCCGAGCGCTGCATTAGACTACTTCAAGTCTAA
FUN_000001 5952 12 5952 5953
FUN_000002 5072 5977 5072 5073
FUN_000003 1219 11062 1219 1220
FUN_000004 7971 12294 7971 7972
FUN_000005 5072 20278 5072 5073
FUN_000006 16585 25363 16585 16586
FUN_000007 1032 41961 1032 1033
FUN_000008 638 43006 638 639
FUN_000009 9245 43657 9245 9246
FUN_000010 1750 52915 1750 17514 Subset Expressed Genes
Only those with at least one read in each sample
4.0.1 Peek at count matrix
head "${count_matrix}" | column -t -s","
echo ""
echo ""
wc -l "${count_matrix}"gene_id RNA-ACR-140 RNA-ACR-145 RNA-ACR-150 RNA-ACR-173 RNA-ACR-178
FUN_035039 553 340 256 485 510
FUN_035038 2486 775 743 1250 1092
FUN_035031 46 6 25 41 29
FUN_035030 183 252 48 78 73
FUN_035033 1519 311 555 990 370
FUN_035032 1764 1297 1035 1763 1360
FUN_035035 601 256 231 521 126
FUN_035034 3644 3770 3137 3225 3322
FUN_035037 5546 4928 3061 4720 3018
44372 ../output/07-Apul-Hisat/Apul-gene_count_matrix.csv4.1 Import count matrix
# Read the data into a data frame
count_matrix_df <- read.csv(count_matrix, header = TRUE)
str(count_matrix_df)'data.frame': 44371 obs. of 6 variables:
$ gene_id : chr "FUN_035039" "FUN_035038" "FUN_035031" "FUN_035030" ...
$ RNA.ACR.140: int 553 2486 46 183 1519 1764 601 3644 5546 1224 ...
$ RNA.ACR.145: int 340 775 6 252 311 1297 256 3770 4928 301 ...
$ RNA.ACR.150: int 256 743 25 48 555 1035 231 3137 3061 511 ...
$ RNA.ACR.173: int 485 1250 41 78 990 1763 521 3225 4720 677 ...
$ RNA.ACR.178: int 510 1092 29 73 370 1360 126 3322 3018 841 ...4.2 Only genes with at least one read per sample
# Filter rows where all values are greater than 0
filtered_count_matrix_df <- count_matrix_df[apply(count_matrix_df > 0, 1, all), ]
str(filtered_count_matrix_df)'data.frame': 19789 obs. of 6 variables:
$ gene_id : chr "FUN_035039" "FUN_035038" "FUN_035031" "FUN_035030" ...
$ RNA.ACR.140: int 553 2486 46 183 1519 1764 601 3644 5546 1224 ...
$ RNA.ACR.145: int 340 775 6 252 311 1297 256 3770 4928 301 ...
$ RNA.ACR.150: int 256 743 25 48 555 1035 231 3137 3061 511 ...
$ RNA.ACR.173: int 485 1250 41 78 990 1763 521 3225 4720 677 ...
$ RNA.ACR.178: int 510 1092 29 73 370 1360 126 3322 3018 841 ...4.3 Subset genes FastA
Only expressed genes
# Get the row names (gene_ids) of the filtered data frame
filtered_gene_ids <- filtered_count_matrix_df$gene_id
fasta <- readDNAStringSet(genes_fasta)
subset_fasta <- fasta[names(fasta) %in% filtered_gene_ids]writeXStringSet(subset_fasta, genes_subset_fasta)4.3.1 Peek at genes subset FastA
head "${genes_subset_fasta}"
echo ""
grep "^>" --count "${genes_subset_fasta}">FUN_000001
ATGATGCCACAAGGGTACAAAAACGCCCTTCCAGGCTATAAAGACCTCTATCTTAGCCAAGCAATCACCGAGGAGGTTCA
CAATGTAAGTTTGTCCTTTTAGCTTGTGCCTGTTTTTCCTAGTTTGTTTGCTTGTTTCTTTCTTTCTTTCTTTCTTTTTA
GTTCTATTTTGTTTCATTTGTGATTCTTCAAGATATTTGGTGGTTACTTGTTCAGTCTAGCGTAGCCAAACTATTTGAAC
ACTACAAGATAATTCGCAAGAAACAGGTTGAAGAAAATTCCATTTCAAGCGAAATCTTATTTCATTTTTTTACATTCTCT
GTAGCTAATATACTGGCAAGTAATTTTATCGCCAACTGAAGGCAAGCACCAAGGACGTTTACATGGTAAGTTTGTCCTTT
TGTTACACAAATTGGGTAGTCTGTTTCCTGGTGTTTTTTTTTTTTTTAATGTTTTAATTCACGATTATTCAGGATATCTA
GGGATGAATTATCTGTTTATTACTCCAACATAATTAGTAAGAAACCAATTGGGAAGAATTCCAATAGAAGCAAAATCTTA
ATCTGTCTTTAAACTACTATGTAGTTAATACTTTAGCGAGTGATCTCTTCTCGGTGGCTATTACCATTTGTTGTGAATGC
TATGAACGAAGAGCACCTTATTTTACTACCCGACATTATTGTAGCCCTGAGTTCCCTTTAGATCTGACGCTGCCATTTCT
197895 BLASTx
5.1 Download SwissProt
cd "${blastdbs_dir}"
curl -O https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gz
mv uniprot_sprot.fasta.gz 20250618-uniprot_sprot.fasta.gz
gunzip --keep 20250618-uniprot_sprot.fasta.gz % Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0 0 88.7M 0 7853 0 0 23582 0 1:05:47 --:--:-- 1:05:47 23511 16 88.7M 16 14.5M 0 0 11.1M 0 0:00:07 0:00:01 0:00:06 11.1M 55 88.7M 55 49.3M 0 0 21.3M 0 0:00:04 0:00:02 0:00:02 21.3M 97 88.7M 97 86.2M 0 0 25.9M 0 0:00:03 0:00:03 --:--:-- 25.9M100 88.7M 100 88.7M 0 0 26.2M 0 0:00:03 0:00:03 --:--:-- 26.2M5.2 Create BLASTdb
cd "${blastdbs_dir}"
"${diamond}" makedb \
--in 20250618-uniprot_sprot.fasta.gz \
--db "${diamond_db}" \
--quiet \
--threads "${threads}"5.3 Run DIAMOND BLASTx
"${diamond}" blastx \
--db "${blastdbs_dir}"/"${diamond_db}.dmnd" \
--query "${genes_subset_fasta}" \
--out "${diamond_output}" \
--outfmt 6 \
--sensitive \
--evalue 1e-10 \
--max-target-seqs 1 \
--block-size 15.0 \
--index-chunks 4 \
--threads "${threads}" \
2> ../output/30.00-Apul-transcriptome-GOslims/diamond-blastx.log
head "${diamond_output}"
echo ""
wc -l "${diamond_output}"FUN_000067 sp|P84227|H32_BOVIN 99.3 136 1 0 1 408 1 136 2.74e-84 250
FUN_000084 sp|P84227|H32_BOVIN 99.3 136 1 0 1 408 1 136 2.74e-84 250
FUN_000090 sp|P84227|H32_BOVIN 99.3 136 1 0 1 408 1 136 2.74e-84 250
FUN_000094 sp|P84227|H32_BOVIN 99.3 136 1 0 1 408 1 136 2.74e-84 250
FUN_000102 sp|P84227|H32_BOVIN 99.3 136 1 0 1 408 1 136 2.74e-84 250
FUN_000122 sp|P35061|H2A_ACRFO 100 125 0 0 711 1085 1 125 1.97e-74 240
FUN_000123 sp|P84227|H32_BOVIN 99.3 136 1 0 1 408 1 136 2.74e-84 250
FUN_000133 sp|P35059|H4_ACRFO 100 103 0 0 312 4 1 103 4.15e-66 197
FUN_000141 sp|P35067|H2B_ACRFO 99.2 125 1 0 1211 837 1 125 8.07e-74 229
FUN_000149 sp|P35067|H2B_ACRFO 98.4 125 2 0 2442 2068 1 125 1.61e-69 228
9141 ../output/30.00-Apul-transcriptome-GOslims/Apulchra-expressed-genes.blastx.outfmt66 GENE ONTOLOGY
6.1 Get gene IDs and SwissProt IDs
awk -F"|" '{print $1"\t"$2}' "${diamond_output}" \
| awk '{print $1"\t"$3}' \
> ../output/30.00-Apul-transcriptome-GOslims/gene-SPIDs.txt
head ../output/30.00-Apul-transcriptome-GOslims/gene-SPIDs.txt
echo ""
echo ""
wc -l ../output/30.00-Apul-transcriptome-GOslims/gene-SPIDs.txtFUN_000067 P84227
FUN_000084 P84227
FUN_000090 P84227
FUN_000094 P84227
FUN_000102 P84227
FUN_000122 P35061
FUN_000123 P84227
FUN_000133 P35059
FUN_000141 P35067
FUN_000149 P35067
9141 ../output/30.00-Apul-transcriptome-GOslims/gene-SPIDs.txt6.2 Get SwissProt IDs
awk -F"|" '{print $2}' "${diamond_output}" \
| sort --unique \
> ../output/30.00-Apul-transcriptome-GOslims/SPIDs.txt
head ../output/30.00-Apul-transcriptome-GOslims/SPIDs.txt
echo ""
echo ""
wc -l ../output/30.00-Apul-transcriptome-GOslims/SPIDs.txtA0A061IR73
A0A072UTP9
A0A075F932
A0A096X8J7
A0A0A2JW91
A0A0C2SRU0
A0A0G2JZ79
A0A0G2K047
A0A0G2K2P5
A0A0G2L7I0
6283 ../output/30.00-Apul-transcriptome-GOslims/SPIDs.txt6.3 Retrieve UniProt records
A difference in number of records could be due to retrieval from only “reviewed” records, while BLAST may have included both “reviewed” and “unreviewed.” SwissProt records
python3 \
../../M-multi-species/code/uniprot-retrieval.py \
../output/30.00-Apul-transcriptome-GOslims/SPIDs.txt \
../output/30.00-Apul-transcriptome-GOslims/
gunzip ../output/30.00-Apul-transcriptome-GOslims/uniprot-retrieval.tsv.gz
echo ""
echo ""
wc -l ../output/30.00-Apul-transcriptome-GOslims/uniprot-retrieval.tsv500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
500 / 500
283 / 283
gzip: ../output/30.00-Apul-transcriptome-GOslims/uniprot-retrieval.tsv already exists; not overwritten
6295 ../output/30.00-Apul-transcriptome-GOslims/uniprot-retrieval.tsv6.4 Map GO to Genes
# Read gene-SPIDs.txt
gene_spids <- read.delim("../output/30.00-Apul-transcriptome-GOslims/gene-SPIDs.txt", header = FALSE, stringsAsFactors = FALSE)
colnames(gene_spids) <- c("GeneID", "Entry")
# Read uniprot-retrieval.tsv
uniprot <- read.delim("../output/30.00-Apul-transcriptome-GOslims/uniprot-retrieval.tsv", header = TRUE, stringsAsFactors = FALSE, check.names = FALSE)
# Merge on Entry
gene_SPID_GOID_merged <- merge(gene_spids, uniprot[, c("Entry", "Gene Ontology IDs")], by = "Entry", all.x = TRUE)
# View result
str(gene_SPID_GOID_merged)'data.frame': 9141 obs. of 3 variables:
$ Entry : chr "A0A061IR73" "A0A072UTP9" "A0A075F932" "A0A096X8J7" ...
$ GeneID : chr "FUN_031929" "FUN_010760" "FUN_005029" "FUN_017765" ...
$ Gene Ontology IDs: chr "GO:0001750; GO:0005524; GO:0005778; GO:0005829; GO:0006625; GO:0016558; GO:0016561; GO:0016562; GO:0016887; GO:"| __truncated__ "GO:0004197; GO:0005615; GO:0005764; GO:0006955; GO:0051603; GO:2001235" "GO:0000149; GO:0001786; GO:0005509; GO:0005543; GO:0005544; GO:0005737; GO:0005886; GO:0008021; GO:0017158; GO:"| __truncated__ "GO:0005452; GO:0005886; GO:0008509; GO:0015701; GO:0051453; GO:0055085; GO:0086001" ...6.4.1 Write merged to file
# Optionally, write to file
write.table(gene_SPID_GOID_merged, "../output/30.00-Apul-transcriptome-GOslims/gene-SPIDs-GOIDs.tsv", sep = "\t", row.names = FALSE, quote = FALSE)6.5 Clean up merged
full.gene.df <- as.data.frame(gene_SPID_GOID_merged)
# Clean whitespace, filter NA/empty rows, select columns, and split GO terms using column name variables
gene.GO.df <- full.gene.df %>%
mutate(!!"Gene Ontology IDs" := str_replace_all(.data[["Gene Ontology IDs"]], "\\s*;\\s*", ";")) %>% # Clean up spaces around ";"
filter(!is.na(.data[["GeneID"]]) & !is.na(.data[["Gene Ontology IDs"]]) & .data[["Gene Ontology IDs"]] != "") %>%
dplyr::select(all_of(c("GeneID", "Gene Ontology IDs")))
str(gene.GO.df)'data.frame': 9011 obs. of 2 variables:
$ GeneID : chr "FUN_031929" "FUN_010760" "FUN_005029" "FUN_017765" ...
$ Gene Ontology IDs: chr "GO:0001750;GO:0005524;GO:0005778;GO:0005829;GO:0006625;GO:0016558;GO:0016561;GO:0016562;GO:0016887;GO:0043335;G"| __truncated__ "GO:0004197;GO:0005615;GO:0005764;GO:0006955;GO:0051603;GO:2001235" "GO:0000149;GO:0001786;GO:0005509;GO:0005543;GO:0005544;GO:0005737;GO:0005886;GO:0008021;GO:0017158;GO:0030154;G"| __truncated__ "GO:0005452;GO:0005886;GO:0008509;GO:0015701;GO:0051453;GO:0055085;GO:0086001" ...6.6 Flatten gene GOID file
flat.gene.GO.df <- gene.GO.df %>% separate_rows(!!sym("Gene Ontology IDs"), sep = ";")
str(flat.gene.GO.df)tibble [109,816 × 2] (S3: tbl_df/tbl/data.frame)
$ GeneID : chr [1:109816] "FUN_031929" "FUN_031929" "FUN_031929" "FUN_031929" ...
$ Gene Ontology IDs: chr [1:109816] "GO:0001750" "GO:0005524" "GO:0005778" "GO:0005829" ...6.7 Group by GOID
grouped.gene.GO.df <- flat.gene.GO.df %>%
group_by(!!sym("Gene Ontology IDs")) %>%
summarise(!!"GeneID" := paste(.data[["GeneID"]], collapse = ","))
str(grouped.gene.GO.df)tibble [12,284 × 2] (S3: tbl_df/tbl/data.frame)
$ Gene Ontology IDs: chr [1:12284] "GO:0000002" "GO:0000009" "GO:0000012" "GO:0000014" ...
$ GeneID : chr [1:12284] "FUN_025331,FUN_001651,FUN_001649,FUN_033608,FUN_009696,FUN_006166,FUN_035449,FUN_006171,FUN_008305,FUN_011990,FUN_016408" "FUN_008599" "FUN_001342,FUN_011804,FUN_008094,FUN_010555" "FUN_027937,FUN_038659,FUN_038683,FUN_038674,FUN_038658,FUN_038643,FUN_038681,FUN_007893,FUN_024354,FUN_041874" ...6.8 Vectorize GOIDs
# Vector of GO IDs
go_ids <- grouped.gene.GO.df[["Gene Ontology IDs"]]
str(go_ids) chr [1:12284] "GO:0000002" "GO:0000009" "GO:0000012" "GO:0000014" ...6.9 Prepare GOslim OBO
# Find GSEAbase installation location
gseabase_location <- find.package("GSEABase")
# Load path to GOslim OBO file
goslim_obo_destintation <- file.path(gseabase_location, "extdata", goslims_obo, fsep = "/")
# Download the GOslim OBO file
download.file(url = goslims_url, destfile = goslim_obo_destintation)
# Loads package files
gseabase_files <- system.file("extdata", goslims_obo, package="GSEABase")6.10 GOslims from OBO
# Create GSEAbase GOCollection using `go_ids`
myCollection <- GOCollection(go_ids)
# Retrieve GOslims from GO OBO file set
slim <- getOBOCollection(gseabase_files)
str(slim)Formal class 'OBOCollection' [package "GSEABase"] with 7 slots
..@ .stanza :'data.frame': 153 obs. of 1 variable:
.. ..$ value: chr [1:153] "Root" "Term" "Term" "Term" ...
..@ .subset :'data.frame': 22 obs. of 1 variable:
.. ..$ value: chr [1:22] "Rhea list of ChEBI terms representing the major species at pH 7.3." "Term not to be used for direct annotation" "Terms planned for obsoletion" "AGR slim" ...
..@ .kv :'data.frame': 2110 obs. of 3 variables:
.. ..$ stanza_id: chr [1:2110] ".__Root__" ".__Root__" ".__Root__" ".__Root__" ...
.. ..$ key : chr [1:2110] "format-version" "data-version" "synonymtypedef" "synonymtypedef" ...
.. ..$ value : chr [1:2110] "1.2" "go/releases/2025-06-01/subsets/goslim_generic.owl" "syngo_official_label \"label approved by the SynGO project\"" "systematic_synonym \"Systematic synonym\" EXACT" ...
..@ evidenceCode: chr [1:26] "EXP" "IDA" "IPI" "IMP" ...
..@ ontology : chr NA
..@ ids : chr [1:141] "GO:0000228" "GO:0000278" "GO:0000910" "GO:0001618" ...
..@ type : chr "OBO"6.11 Biological Process GOslims
# Retrieve Biological Process (BP) GOslims
slimdf <- goSlim(myCollection, slim, "BP", verbose)
str(slimdf)'data.frame': 72 obs. of 3 variables:
$ Count : int 90 18 20 430 63 75 18 3 90 51 ...
$ Percent: num 1.11 0.222 0.247 5.305 0.777 ...
$ Term : chr "mitotic cell cycle" "cytokinesis" "cytoplasmic translation" "immune system process" ...6.12 Map GO to GOslims
# List of GOslims and all GO IDs from `go_ids`
gomap <- as.list(GOBPOFFSPRING[rownames(slimdf)])
# Maps `go_ids` to matching GOslims
mapped <- lapply(gomap, intersect, ids(myCollection))
# Append all mapped GO IDs to `slimdf`
# `sapply` needed to apply paste() to create semi-colon delimited values
slimdf$GO.IDs <- sapply(lapply(gomap, intersect, ids(myCollection)), paste, collapse=";")
# Remove "character(0) string from "GO.IDs" column
slimdf$GO.IDs[slimdf$GO.IDs == "character(0)"] <- ""
# Add self-matching GOIDs to "GO.IDs" column, if not present
for (go_id in go_ids) {
# Check if the go_id is present in the row names
if (go_id %in% rownames(slimdf)) {
# Check if the go_id is not present in the GO.IDs column
# Also removes white space "trimws()" and converts all to upper case to handle
# any weird, "invisible" formatting issues.
if (!go_id %in% trimws(toupper(strsplit(slimdf[go_id, "GO.IDs"], ";")[[1]]))) {
# Append the go_id to the GO.IDs column with a semi-colon separator
if (length(slimdf$GO.IDs) > 0 && nchar(slimdf$GO.IDs[nrow(slimdf)]) > 0) {
slimdf[go_id, "GO.IDs"] <- paste0(slimdf[go_id, "GO.IDs"], "; ", go_id)
} else {
slimdf[go_id, "GO.IDs"] <- go_id
}
}
}
}
str(slimdf)'data.frame': 72 obs. of 4 variables:
$ Count : int 90 18 20 430 63 75 18 3 90 51 ...
$ Percent: num 1.11 0.222 0.247 5.305 0.777 ...
$ Term : chr "mitotic cell cycle" "cytokinesis" "cytoplasmic translation" "immune system process" ...
$ GO.IDs : chr "GO:0000022;GO:0000070;GO:0000082;GO:0000086;GO:0000132;GO:0000281;GO:0007052;GO:0007064;GO:0007076;GO:0007079;G"| __truncated__ "GO:0000281;GO:0000915;GO:0007110;GO:0007111;GO:0007112;GO:0032465;GO:0032466;GO:0032467;GO:0036089;GO:0036090;G"| __truncated__ "GO:0001731;GO:0001732;GO:0002182;GO:0002183;GO:0002188;GO:0002191;GO:0017183;GO:0140018;GO:0140708;GO:1900249;G"| __truncated__ "GO:0001771;GO:0001774;GO:0001776;GO:0001779;GO:0001780;GO:0001782;GO:0001787;GO:0001805;GO:0001812;GO:0001867;G"| __truncated__ ...6.13 Flatten GOslims
# "Flatten" file so each row is single GO ID with corresponding GOslim
# rownames_to_column needed to retain row name info
slimdf_separated <- as.data.frame(slimdf %>%
rownames_to_column('GOslim') %>%
separate_rows(GO.IDs, sep = ";"))
# Group by unique GO ID
grouped_slimdf <- slimdf_separated %>%
filter(!is.na(GO.IDs) & GO.IDs != "") %>%
group_by(GO.IDs) %>%
summarize(GOslim = paste(GOslim, collapse = ";"),
Term = paste(Term, collapse = ";"))
str(grouped_slimdf)tibble [5,628 × 3] (S3: tbl_df/tbl/data.frame)
$ GO.IDs: chr [1:5628] " GO:0000278" " GO:0002181" " GO:0002376" " GO:0003014" ...
$ GOslim: chr [1:5628] "GO:0000278" "GO:0002181" "GO:0002376" "GO:0003014" ...
$ Term : chr [1:5628] "mitotic cell cycle" "cytoplasmic translation" "immune system process" "renal system process" ...6.14 Counts of GOslims
slimdf.sorted <- slimdf %>% arrange(desc(Count))
slim.count.df <- slimdf.sorted %>%
dplyr::select(Term, Count, Percent)
str(slim.count.df)'data.frame': 72 obs. of 3 variables:
$ Term : chr "anatomical structure development" "cell differentiation" "signaling" "immune system process" ...
$ Count : int 1768 739 728 430 285 254 251 204 199 182 ...
$ Percent: num 21.81 9.12 8.98 5.3 3.52 ...6.14.1 Write GOslims to file
Need to create a column name for GOslimIDs from data frame rownames.
# Create header vector
header <- c("GOslimID", colnames(slim.count.df))
# Write header to file
writeLines(paste(header, collapse = "\t"), "../output/30.00-Apul-transcriptome-GOslims/GOslim-counts.tsv")
# Append data frame contents to existing file, which contains header info
write.table(
slim.count.df,
"../output/30.00-Apul-transcriptome-GOslims/GOslim-counts.tsv",
sep = "\t",
row.names = TRUE,
quote = FALSE,
col.names = FALSE,
append = TRUE
)RESULTS
Multispecies barplots were created of the top 10 GOslims with the highest GO term counts.
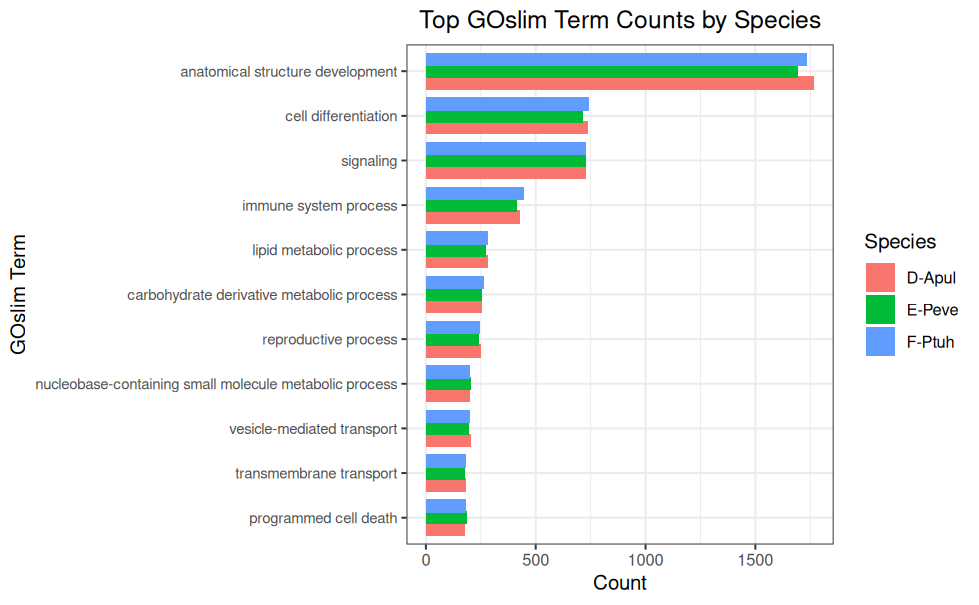
Plots were generated with the following code:
# Define file paths and species names
files <- list(
"D-Apul" = "~/gitrepos/urol-e5/deep-dive-expression/D-Apul/output/30.00-Apul-transcriptome-GOslims/GOslim-counts.tsv",
"E-Peve" = "~/gitrepos/urol-e5/deep-dive-expression/E-Peve/output/30.00-Peve-transcriptome-GOslims/GOslim-counts.tsv",
"F-Ptuh" = "~/gitrepos/urol-e5/deep-dive-expression/F-Ptuh/output/30.00-Ptua-transcriptome-GOslims/GOslim-counts.tsv"
)
# Read and combine data
goslim_data <- map2_dfr(
files,
names(files),
~ read_tsv(.x, show_col_types = FALSE) %>%
mutate(Species = .y)
)
# Find the top 10 Terms by Count for each species
top_terms <- goslim_data %>%
group_by(Species) %>%
slice_max(order_by = Count, n = 10, with_ties = FALSE) %>%
ungroup() %>%
distinct(Term)
# Filter the data to only include these Terms
goslim_top <- goslim_data %>%
filter(Term %in% top_terms$Term)
# Plot: Term on Y, Count on X
ggplot(goslim_top, aes(y = fct_reorder(Term, Count), x = Count, fill = Species)) +
geom_bar(stat = "identity", position = position_dodge(width = 0.8)) +
labs(
title = "Top GOslim Term Counts by Species",
y = "GOslim Term",
x = "Count"
) +
theme_bw() +
theme(
axis.text.y = element_text(size = 8),
plot.title = element_text(hjust = 0.5)
)