Previously trimmed the first 39 bases of sequence from reads from the BS-Seq data in an attempt to improve our ability to map the reads back to the C.gigas genome. However, Mac (and Steven) noticed that the last ~10 bases of all the reads showed a steady increase in the %G, suggesting some sort of bias (maybe adaptor??):
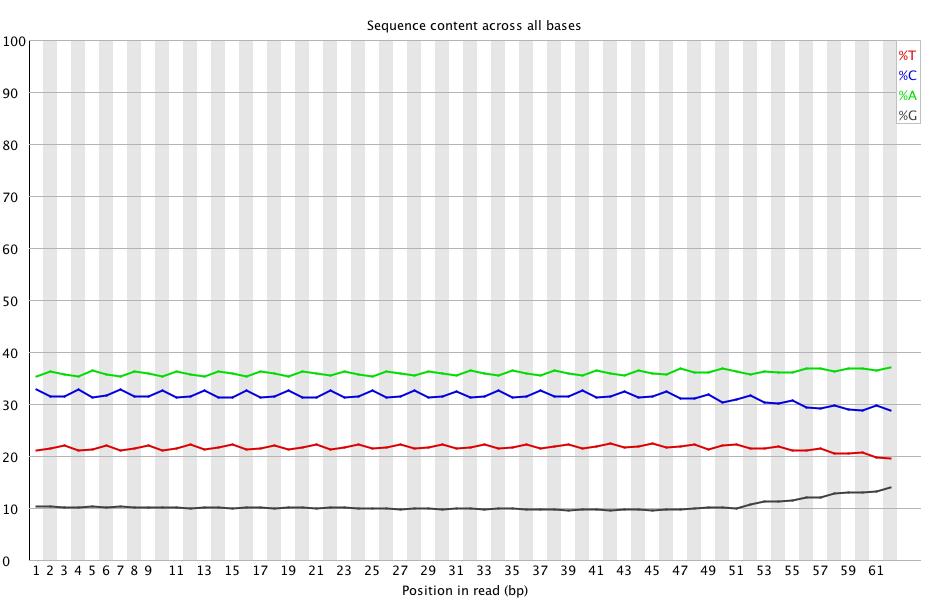 (http://eagle.fish.washington.edu/Arabidopsis/20150506_trimmed_2212_lane2_CTTGTA_L002_R1_001_fastqc/Images/per_base_sequence_content.png)
(http://eagle.fish.washington.edu/Arabidopsis/20150506_trimmed_2212_lane2_CTTGTA_L002_R1_001_fastqc/Images/per_base_sequence_content.png)
Although I didn’t mention this previously, the figure above also shows an odd “waves” pattern that repeats in all bases except for G. Not sure what to think of that…
Quick summary of actions taken (specifics are available in Jupyter notebook below):
Trim first 39 bases from all reads in all raw sequencing files.
Trim last 10 bases from all reads in raw sequencing files
Concatenate the two sets of reads (400ppm and 1000ppm treatments) into single FASTQ files for Steven to work with.
Raw sequencing files:
Notebook Viewer: 20150521_Cgigas_larvae_OA_Trimmomatic_FASTQC
Jupyter (IPython) notebook: 20150521_Cgigas_larvae_OA_Trimmomatic_FASTQC.ipynb
Output files
Trimmed, concatenated FASTQ files 20150521_trimmed_2212_lane2_400ppm_GCCAAT.fastq.gz 20150521_trimmed_2212_lane2_1000ppm_CTTGTA.fastq.gz
FASTQC files 20150521_trimmed_2212_lane2_400ppm_GCCAAT_fastqc.html 20150521_trimmed_2212_lane2_400ppm_GCCAAT_fastqc.zip
20150521_trimmed_2212_lane2_1000ppm_CTTGTA_fastqc.html 20150521_trimmed_2212_lane2_1000ppm_CTTGTA_fastqc.zip
Example of FASTQC analysis pre-trim:
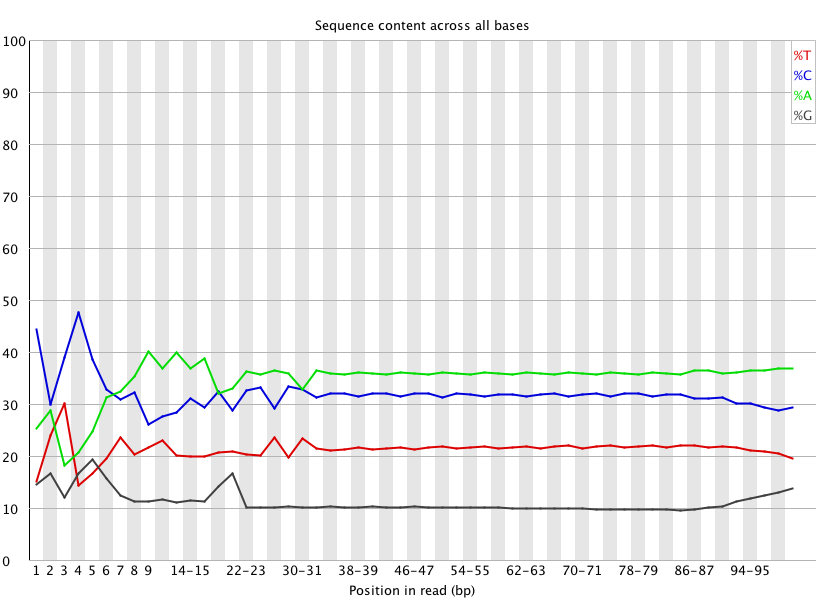 (http://eagle.fish.washington.edu/Arabidopsis/20150414_trimmed_2212_lane2_CTTGTA_L002_R1_001_fastqc/Images/per_base_sequence_content.png)
(http://eagle.fish.washington.edu/Arabidopsis/20150414_trimmed_2212_lane2_CTTGTA_L002_R1_001_fastqc/Images/per_base_sequence_content.png)
Example FASTQC post-trim (from 400ppm data):
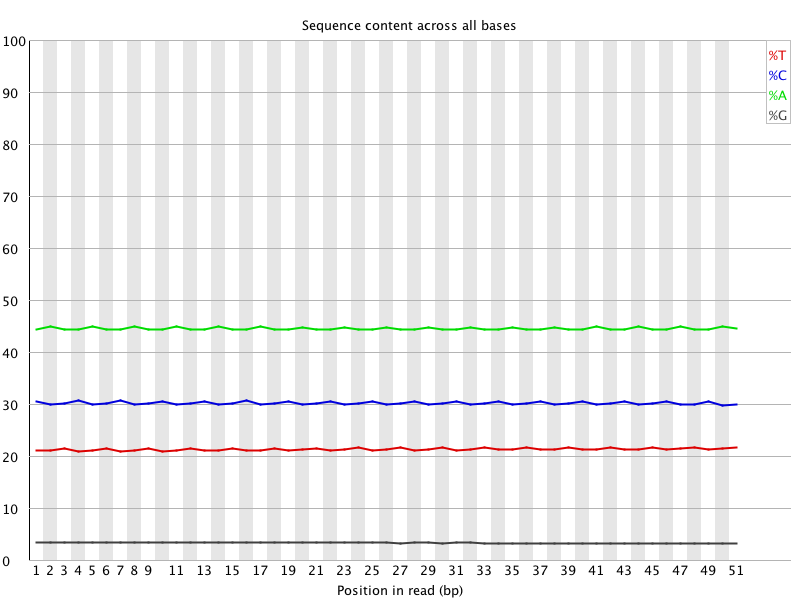 (http://eagle.fish.washington.edu/Arabidopsis/20150521_trimmed_2212_lane2_400ppm_GCCAAT_fastqc/Images/per_base_sequence_content.png)
(http://eagle.fish.washington.edu/Arabidopsis/20150521_trimmed_2212_lane2_400ppm_GCCAAT_fastqc/Images/per_base_sequence_content.png)
Trimming has removed the intended bad stuff (inconsistent sequence in the first 39 bases and rise in %G in the last 10 bases). Sequences are ready for further analysis for Steven.
However, we still see the “waves” pattern with the T, A and C. Additionally, we still don’t know what caused the weird inconsistencies, nor what sequence is contained therein that might be leading to that. Will contact the sequencing facility to see if they have any insight.