Needed to assess the integrity of the newest gDNA isolated for the two genome sequencing projects: Geoduck gDNA from earlier today and Olympia oyster gDNA from 20151003.
Also needed to assess the integrity of the gDNA of ethanol-preserved Olympia oyster mantle tissue from Jake’s reciprocal transplant experiment, isolated on 20151003: samples NF1A & SN49A.
Ran samples on a 0.8% agarose, 1X modified TAE gel.
Loaded 1μL (~300ng) of the geoduck, Oly and NF1A samples.
Loaded 2μL (~100ng) of the SN49A sample.
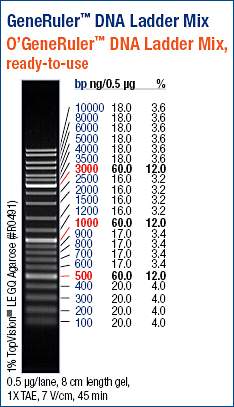
Used 5μL of ladder.
Results:
 (https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
(https://raw.githubusercontent.com/sr320/LabDocs/master/protocols/Commercial_Protocols/ThermoFisher_OgeneRuler_DNA_Ladder_Mix_F100439.jpg)
 (http://eagle.fish.washington.edu/Arabidopsis/20151005_gDNA_gel_annotated.jpg)
(http://eagle.fish.washington.edu/Arabidopsis/20151005_gDNA_gel_annotated.jpg)
Genome Sequencing Samples
The geoduck and the Oly samples look good. Intact, high molecular weight band. My only concern is the noticeable difference in band intensities between these two. Both samples should be ~300ng/μL, based on the NanoDrop1000 readings. However, it’s evident that the concentrations of these two samples differ greatly. Additionally, we can use the ladder to gauge the concentrations of the samples, since I loaded 0.5μg of the ladder, which is the quantity referenced on the ladder guide above.
It would appear that the geoduck sample concentration is closer to 60ng/μL (band intensity is similar to that of the 500, 1000, & 3000bp markers), as opposed to the 292ng/μL that the NanoDrop1000 indicated.
The Oly sample appears to have even less and appears less intense than the lowest concentration bands on the marker (16.0ng/μL). That’s not even remotely close to the 331ng/μL measured by the NanoDrop1000.
It’s difficult to say why this might be, as both samples were RNased and neither of them show extensive smearing (both of those factors would contribute to inflated spec readings).
Regardless, will ship them off to BGI to supplement the previous gDNA for this project.
Ethanol-Preserved Samples
Both samples show extensive smearing and no high molecular weight band, indicating they are both completely degraded. This is a very bad result for this project, as the tissue in this group is/was a bit of grasping at straws to obtain some intact DNA to use for the RAD-seq that we intend to pursue.