INTRO
After a previous attempt at amplifying the C1q gene in bisulfite-converted DNA failed, we decided we should test out a standard PCR on untreated liver gDNA. I designed PCR primers for the C1Q gene on 20240916, I decided to evaluate Salvelinus namaycush gDNA integrity isolated on 20240712 to see if the DNA was degraded (or not).
MATERIALS & METHODS
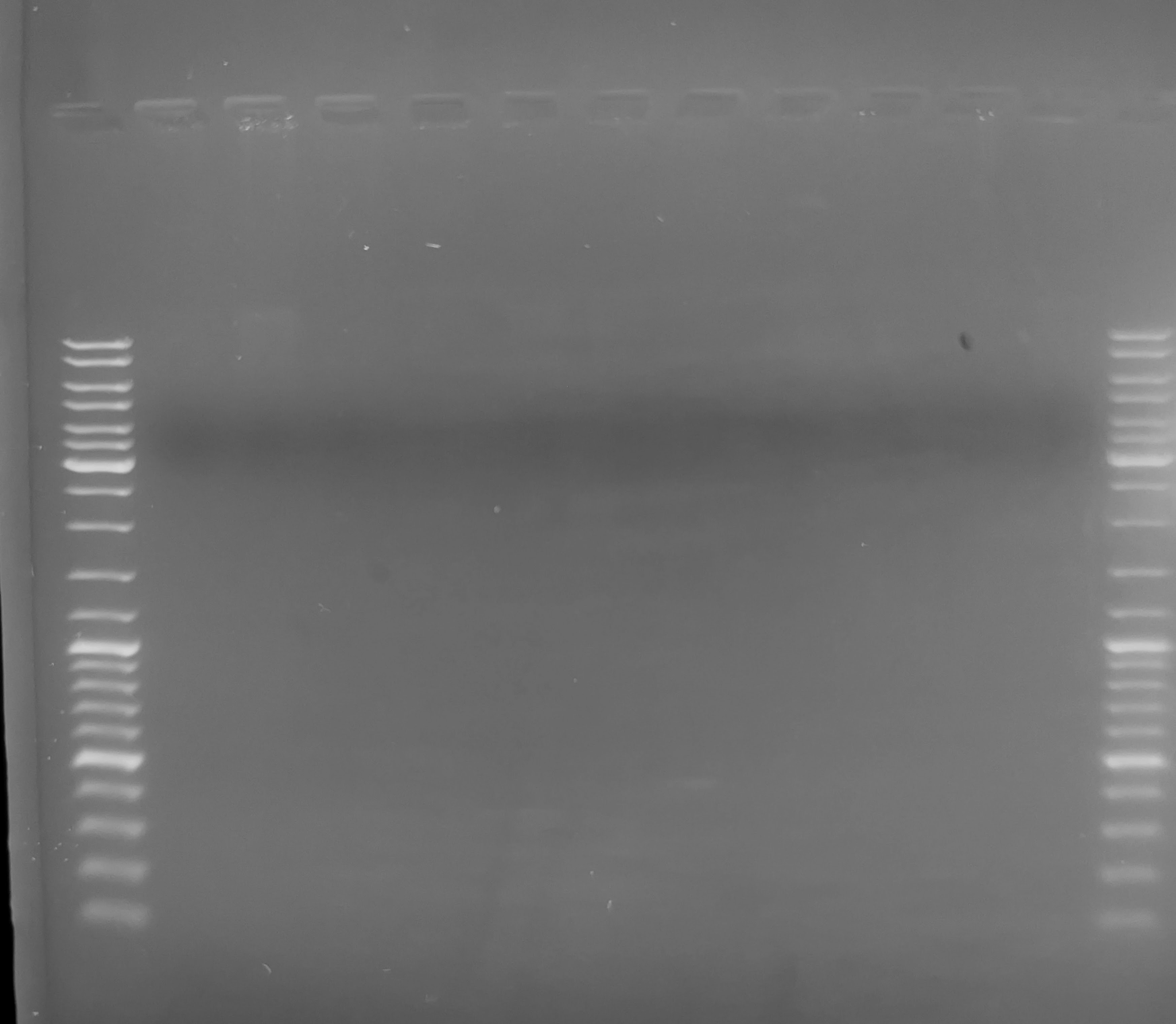
Samples were run on 1.0% agarose, low-TAE gel with ethidium bromide. I prepared the DNA for loading by combining 0.5uL of each sample with 19.5uL of 1x TE, and 4uL of 6x Orange DNA Loading Dye (Thermo Scientific). The gel was run @ 107V for ~45mins and then imaged.
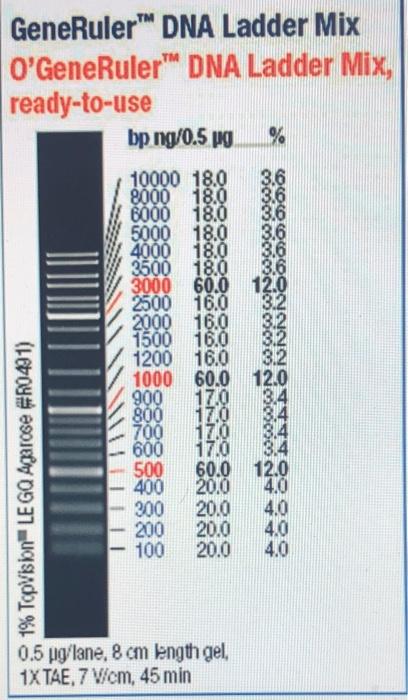
O’GeneRuler DNA Ladder Mix, Ready-to-Use 100-10,00bp (Thermo Scientific) was used (5uL) for size reference:
RESULTS
Well, there’s no visible DNA on the gel. This suggests that an insufficient amount of DNA was loaded. The reason this is the best explanation is that even if the DNA were degraded, we’d still expect to see smearing, which would reflect degradation. Instead, we don’t see anything.
DISCUSSION
Not sure exactly why this might be, as quantification would indicate the use of 0.5uL of each sample should yield more than enough to visualize on a gel (~100ng is usually sufficient for visualization) (Notebook entry).