For the first task you will take an unknown multi-fasta file and annotate it using blast. You are welcome to do this in terminal, Rstudio, or jupyter. My recommendation, and how I will demonstrate is using Rmarkdown. Once you have have your project structured, we will download software, databases, a fasta file and run the code.
This is product offers a workflow to take a few thousand unidentified sequences and provide a better understanding of what genes are present. This will be accomplished through using Blast and protein sequenes from UniProt/Swiss-prot.
A few weeks ago I perfected software installation, so I will not demonstrate that here. Please see this notebook for more.
Database Creation
Obtain Fasta (UniProt/Swiss-Prot)
This is from here picur reviewe sequences I named based on the identify of the database given
current_time <-format(Sys.time(), "%B %d, %Y %H:%M:%S")cat("current date and time is ", current_time)
cd ../datacurl-O https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gzmv uniprot_sprot.fasta.gz uniprot_sprot_r2023_04.fasta.gzgunzip-k uniprot_sprot_r2023_04.fasta.gz
echo"How many sequences are there?"grep-c">" ../data/Ab_4denovo_CLC6_a.fa
# Read FASTA filefasta_file <-"../data/Ab_4denovo_CLC6_a.fa"# Replace with the name of your FASTA filesequences <-readDNAStringSet(fasta_file)# Calculate sequence lengthssequence_lengths <-width(sequences)# Create a data framesequence_lengths_df <-data.frame(Length = sequence_lengths)# Plot histogram using ggplot2ggplot(sequence_lengths_df, aes(x = Length)) +geom_histogram(binwidth =1, color ="grey", fill ="blue", alpha =0.75) +labs(title ="Histogram of Sequence Lengths",x ="Sequence Length",y ="Frequency") +theme_minimal()
# Read FASTA filefasta_file <-"../data/Ab_4denovo_CLC6_a.fa"sequences <-readDNAStringSet(fasta_file)# Calculate base compositionbase_composition <-alphabetFrequency(sequences, baseOnly =TRUE)# Convert to data frame and reshape for ggplot2base_composition_df <-as.data.frame(base_composition)base_composition_df$ID <-rownames(base_composition_df)base_composition_melted <- reshape2::melt(base_composition_df, id.vars ="ID", variable.name ="Base", value.name ="Count")# Plot base composition bar chart using ggplot2ggplot(base_composition_melted, aes(x = Base, y = Count, fill = Base)) +geom_bar(stat ="identity", position ="dodge", color ="black") +labs(title ="Base Composition",x ="Base",y ="Count") +theme_minimal() +scale_fill_manual(values =c("A"="green", "C"="blue", "G"="yellow", "T"="red"))
# Read FASTA filefasta_file <-"../data/Ab_4denovo_CLC6_a.fa"sequences <-readDNAStringSet(fasta_file)# Count CG motifs in each sequencecount_cg_motifs <-function(sequence) { cg_motif <-"CG"return(length(gregexpr(cg_motif, sequence, fixed =TRUE)[[1]]))}cg_motifs_counts <-sapply(sequences, count_cg_motifs)# Create a data framecg_motifs_counts_df <-data.frame(CG_Count = cg_motifs_counts)# Plot CG motifs distribution using ggplot2ggplot(cg_motifs_counts_df, aes(x = CG_Count)) +geom_histogram(binwidth =1, color ="black", fill ="blue", alpha =0.75) +labs(title ="Distribution of CG Motifs",x ="Number of CG Motifs",y ="Frequency") +theme_minimal()
# Read datasetdataset <-read.csv("../output/blast_annot_go.tab", sep ='\t') # Replace with the path to your dataset# Select the column of interestcolumn_name <-"Organism"# Replace with the name of the column of interestcolumn_data <- dataset[[column_name]]# Count the occurrences of the strings in the columnstring_counts <-table(column_data)# Convert to a data frame, sort by count, and select the top 10string_counts_df <-as.data.frame(string_counts)colnames(string_counts_df) <-c("String", "Count")string_counts_df <- string_counts_df[order(string_counts_df$Count, decreasing =TRUE), ]top_10_strings <-head(string_counts_df, n =10)# Plot the top 10 most common strings using ggplot2ggplot(top_10_strings, aes(x =reorder(String, -Count), y = Count, fill = String)) +geom_bar(stat ="identity", position ="dodge", color ="black") +labs(title ="Top 10 Species hits",x = column_name,y ="Count") +theme_minimal() +theme(legend.position ="none") +coord_flip()
data <-read.csv("../output/blast_annot_go.tab", sep ='\t')# Rename the `Gene.Ontology..biological.process.` column to `Biological_Process`colnames(data)[colnames(data) =="Gene.Ontology..biological.process."] <-"Biological_Process"# Separate the `Biological_Process` column into individual biological processesdata_separated <-unlist(strsplit(data$Biological_Process, split =";"))# Trim whitespace from the biological processesdata_separated <-gsub("^\\s+|\\s+$", "", data_separated)# Count the occurrences of each biological processprocess_counts <-table(data_separated)process_counts <-data.frame(Biological_Process =names(process_counts), Count =as.integer(process_counts))process_counts <- process_counts[order(-process_counts$Count), ]# Select the 20 most predominant biological processestop_20_processes <- process_counts[1:20, ]# Create a color palette for the barsbar_colors <-rainbow(nrow(top_20_processes))# Create a staggered vertical bar plot with different colors for each barbarplot(top_20_processes$Count, names.arg =rep("", nrow(top_20_processes)), col = bar_colors,ylim =c(0, max(top_20_processes$Count) *1.25),main ="Occurrences of the 20 Most Predominant Biological Processes", xlab ="Biological Process", ylab ="Count")# Create a separate plot for the legendpng("../output/GOlegend.png", width =800, height =600)par(mar =c(0, 0, 0, 0))plot.new()legend("center", legend = top_20_processes$Biological_Process, fill = bar_colors, cex =1, title ="Biological Processes")dev.off()
knitr::include_graphics("../output/GOlegend.png")
Case Study - Oyster
Navigating Annotation
The following is a stepwise example or annotation of a gene set using UniProt::Swiss-Prot (reviewed) such that Gene Ontology terms can be associated with each gene.
In this following chunk where the fasta file is downloaded the release is noted and the file name is modified accordingly.
cd DRAFT_Funct_Enrich/annotcurl-O https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gzmv uniprot_sprot.fasta.gz uniprot_sprot_r2023_02.fasta.gzgunzip-k uniprot_sprot_r2023_02.fasta.gz
In a majority of cases you will want to annotate a gene set to get gene ontology information. If you are creating your own genome or transcriptome it should be rather straightforward to know what file to annotate. If using a widely studied system where there are publically available resources, it is advisable to use those as this is the best way to facilitate integration of data sets. In this case study we will be considering the Eastern oyster, (Crassostrea virginica) for which there is data at NCBI and Ensembl Metazoa. At NCBI there is both a GenBank and RefSeq assembly available.
In order to know which of the numerous fasta files should annotated with gene ontology information one should think downstream (or look to files already generated) to the identifiers in genesets that would be subject to functional enrichment tests.
The resulting fpkm count matrix for our case study is from an experiment where male and female oysters where exposed to low pH (and control) conditions. The count matrix is accessible here (csv). Hisat2/Stringtie was used to generate the count matrix with GCF_002022765.2_C_virginica-3.0_genomic.gff formatting thus responsible for gene naming. Specifically the naming format is as follows gene-LOC111099033,gene-LOC111099034,gene-LOC111099035.
The following fasta was selected for annotation: GCF_002022765.2_C_virginica-3.0_translated_cds.faa.gz
cd DRAFT_Funct_Enrich/annotcurl-O https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/022/765/GCF_002022765.2_C_virginica-3.0/GCF_002022765.2_C_virginica-3.0_translated_cds.faa.gzgunzip-k GCF_002022765.2_C_virginica-3.0_translated_cds.faa.gz
head-2 DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.faaecho"number of sequences"grep-c">" DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.faa
Now we can take the two data frames: A) blast output of taking protein fasta and comparing to uniprot_swiss-prot and B) a tabular version of same fasta file that has ID numbers of importance. Note this importance was determined based on what we want to use down stream.
g.spid <- left_join(blast, cdsftab, by = "V1") - This line is using the left_join() function from dplyr to merge the blast and cdsftab datasets by the column “V1”. A left join retains all the rows in the blast data frame and appends the matching rows in the cdsftab data frame. If there is no match, the result is NA. The result of this operation is assigned to the g.spid object.
mutate(gene = str_extract(V2.y, "(?<=\\[gene=)\\w+")) - This line is using the mutate() function from dplyr to add a new column called “gene” to the data frame. The new column is created by extracting substrings from the “V2.y” column based on the given regular expression pattern "(?<=\\[gene=)\\w+". This regular expression matches and extracts any word (sequence of word characters, i.e., alphanumeric and underscore) that comes after “[gene=”.
select(gene, V11, V2.x) - This line is using the select() function from dplyr to keep only the specified columns (“gene”, “V11”, and “V2.x”) in the data frame.
mutate(SPID = str_extract(V2.x, "(?<=\\|)[^\\|]*(?=\\|)")) - Again, the mutate() function is used to add another new column named “SPID”. This column is created by extracting substrings from the “V2.x” column. The regular expression "(?<=\\|)[^\\|]*(?=\\|)" is designed to extract any character(s) that is/are surrounded by “|” (pipe symbol). This is a common format for delimited strings.
distinct(gene, SPID, .keep_all = TRUE) - This line is using the distinct() function from dplyr to remove duplicate rows based on the “gene” and “SPID” columns. The .keep_all = TRUE argument means that all other columns are also kept in the result, not just the “gene” and “SPID” columns.
The resulting g.spid data frame should have unique rows with respect to the “gene” and “SPID” columns, and it should contain these two new columns, “gene” and “SPID”, extracted from the original data based on specific string patterns.
With a list of matching Swiss-Prot IDs, (technically UniProt Accession number) we can go back to https://www.uniprot.org and grab corresponding GO terms. This can be done via a web or using Python API.
Using Web
Using ID Mapping
id
finished
Now will customize columns to get GO IDs.
custcol
head-2 DRAFT_Funct_Enrich/annot/uniprotGO.tab
Finally we can join table to get “LOCIDs” the notation for our DEGs, with GO terms.
go <-read.csv("DRAFT_Funct_Enrich/annot/uniprotGO.tab", sep ='\t', header =TRUE, row.names=NULL)
---title: "NCBI Blast"subtitle: "Taking a set of unknown sequence files and annotating them"format: html: code-fold: false code-tools: true code-copy: true highlight-style: github code-overflow: wrap---# TLDR (basics)``` /home/shared/ncbi-blast-2.11.0+/bin/makeblastdb \-in ../data/uniprot_sprot_r2023_01.fasta \-dbtype prot \-out ../blastdb/uniprot_sprot_r2023_01`````` /home/shared/ncbi-blast-2.11.0+/bin/blastx \-query ../data/Ab_4denovo_CLC6_a.fa \-db ../blastdb/uniprot_sprot_r2023_01 \-out ../output/Ab_4-uniprot_blastx.tab \-evalue 1E-20 \-num_threads 20 \-max_target_seqs 1 \-outfmt 6```# 546 TutorialFor the first task you will take an unknown multi-fasta file and annotate it using blast. You are welcome to do this in terminal, Rstudio, or jupyter. My recommendation, and how I will demonstrate is using Rmarkdown. Once you have have your project structured, we will download software, databases, a fasta file and run the code.```{r setup, include = FALSE}library(knitr)library(tidyverse)library(kableExtra)library(DT)#library(Biostrings)knitr::opts_chunk$set(echo =TRUE, # Display code chunkseval =FALSE, # Evaluate code chunkswarning =FALSE, # Hide warningsmessage =FALSE, # Hide messagesfig.width =6, # Set plot width in inchesfig.height =4, # Set plot height in inchesfig.align ="center"# Align plots to the center)```This is product offers a workflow to take a few thousand unidentified sequences and provide a better understanding of what genes are present. This will be accomplished through using Blast and protein sequenes from UniProt/Swiss-prot.------------------------------------------------------------------------A few weeks ago I perfected software installation, so I will not demonstrate that here. Please see this notebook for more.## Database Creation### Obtain Fasta (UniProt/Swiss-Prot)This is from here picur reviewe sequences I named based on the identify of the database given```{r time, eval=FALSE, echo=TRUE}current_time <-format(Sys.time(), "%B %d, %Y %H:%M:%S")cat("current date and time is ", current_time)``````{r download-data, engine='bash'}cd ../datacurl -O https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gzmv uniprot_sprot.fasta.gz uniprot_sprot_r2023_04.fasta.gzgunzip -k uniprot_sprot_r2023_04.fasta.gz```### Making the database```{r make-blastdb, engine='bash'}mkdir ../blastdb/home/shared/ncbi-blast-2.11.0+/bin/makeblastdb \-in ../data/uniprot_sprot_r2023_01.fasta \-dbtype prot \-out ../blastdb/uniprot_sprot_r2023_01```## Getting the query fasta file```{r download-query, engine='bash', eval=FALSE}curl https://eagle.fish.washington.edu/cnidarian/Ab_4denovo_CLC6_a.fa \-k \> ../data/Ab_4denovo_CLC6_a.fa```Exploring what fasta file```{r view-query, eval = FALSE, engine='bash'}head -3 ../data/Ab_4denovo_CLC6_a.fa``````{r view2-query, engine='bash', eval=FALSE}echo "How many sequences are there?"grep -c ">" ../data/Ab_4denovo_CLC6_a.fa``````{r histogram, eval=FALSE}# Read FASTA filefasta_file <-"../data/Ab_4denovo_CLC6_a.fa"# Replace with the name of your FASTA filesequences <-readDNAStringSet(fasta_file)# Calculate sequence lengthssequence_lengths <-width(sequences)# Create a data framesequence_lengths_df <-data.frame(Length = sequence_lengths)# Plot histogram using ggplot2ggplot(sequence_lengths_df, aes(x = Length)) +geom_histogram(binwidth =1, color ="grey", fill ="blue", alpha =0.75) +labs(title ="Histogram of Sequence Lengths",x ="Sequence Length",y ="Frequency") +theme_minimal()``````{r ACGT, eval=FALSE}# Read FASTA filefasta_file <-"../data/Ab_4denovo_CLC6_a.fa"sequences <-readDNAStringSet(fasta_file)# Calculate base compositionbase_composition <-alphabetFrequency(sequences, baseOnly =TRUE)# Convert to data frame and reshape for ggplot2base_composition_df <-as.data.frame(base_composition)base_composition_df$ID <-rownames(base_composition_df)base_composition_melted <- reshape2::melt(base_composition_df, id.vars ="ID", variable.name ="Base", value.name ="Count")# Plot base composition bar chart using ggplot2ggplot(base_composition_melted, aes(x = Base, y = Count, fill = Base)) +geom_bar(stat ="identity", position ="dodge", color ="black") +labs(title ="Base Composition",x ="Base",y ="Count") +theme_minimal() +scale_fill_manual(values =c("A"="green", "C"="blue", "G"="yellow", "T"="red"))``````{r cg, eval=FALSE}# Read FASTA filefasta_file <-"../data/Ab_4denovo_CLC6_a.fa"sequences <-readDNAStringSet(fasta_file)# Count CG motifs in each sequencecount_cg_motifs <-function(sequence) { cg_motif <-"CG"return(length(gregexpr(cg_motif, sequence, fixed =TRUE)[[1]]))}cg_motifs_counts <-sapply(sequences, count_cg_motifs)# Create a data framecg_motifs_counts_df <-data.frame(CG_Count = cg_motifs_counts)# Plot CG motifs distribution using ggplot2ggplot(cg_motifs_counts_df, aes(x = CG_Count)) +geom_histogram(binwidth =1, color ="black", fill ="blue", alpha =0.75) +labs(title ="Distribution of CG Motifs",x ="Number of CG Motifs",y ="Frequency") +theme_minimal()```## Running Blastx```{r blastx, engine='bash'}~/applications/ncbi-blast-2.13.0+/bin/blastx \-query ../data/Ab_4denovo_CLC6_a.fa \-db ../blastdb/uniprot_sprot_r2023_01 \-out ../output/Ab_4-uniprot_blastx.tab \-evalue 1E-20 \-num_threads 20 \-max_target_seqs 1 \-outfmt 6``````{r blast-look, engine='bash'}head -2 ../output/Ab_4-uniprot_blastx.tab``````{r blast-look2, engine='bash'}echo "Number of lines in output"wc -l ../output/Ab_4-uniprot_blastx.tab```## Joining Blast table with annoations.### Prepping Blast table for easy join```{r separate, engine='bash'}tr '|''\t'< ../output/Ab_4-uniprot_blastx.tab \> ../output/Ab_4-uniprot_blastx_sep.tabhead -1 ../output/Ab_4-uniprot_blastx_sep.tab```## Could do some cool stuff in R here reading in table```{r read-data}bltabl <-read.csv("../output/Ab_4-uniprot_blastx_sep.tab", sep ='\t', header =FALSE)spgo <-read.csv("https://gannet.fish.washington.edu/seashell/snaps/uniprot_table_r2023_01.tab", sep ='\t', header =TRUE)``````{r}datatable(head(bltabl), options =list(scrollX =TRUE, scrollY ="400px", scrollCollapse =TRUE, paging =FALSE))``````{r spgo-table}datatable(head(spgo), options =list(scrollX =TRUE, scrollY ="400px", scrollCollapse =TRUE, paging =FALSE))``````{r see}datatable(left_join(bltabl, spgo, by =c("V3"="Entry")) %>%select(V1, V3, V13, Protein.names, Organism, Gene.Ontology..biological.process., Gene.Ontology.IDs) %>%mutate(V1 =str_replace_all(V1, pattern ="solid0078_20110412_FRAG_BC_WHITE_WHITE_F3_QV_SE_trimmed", replacement ="Ab")))``````{r join}annot_tab <-left_join(bltabl, spgo, by =c("V3"="Entry")) %>%select(V1, V3, V13, Protein.names, Organism, Gene.Ontology..biological.process., Gene.Ontology.IDs) %>%mutate(V1 =str_replace_all(V1, pattern ="solid0078_20110412_FRAG_BC_WHITE_WHITE_F3_QV_SE_trimmed", replacement ="Ab"))``````{r}# Read datasetdataset <-read.csv("../output/blast_annot_go.tab", sep ='\t') # Replace with the path to your dataset# Select the column of interestcolumn_name <-"Organism"# Replace with the name of the column of interestcolumn_data <- dataset[[column_name]]# Count the occurrences of the strings in the columnstring_counts <-table(column_data)# Convert to a data frame, sort by count, and select the top 10string_counts_df <-as.data.frame(string_counts)colnames(string_counts_df) <-c("String", "Count")string_counts_df <- string_counts_df[order(string_counts_df$Count, decreasing =TRUE), ]top_10_strings <-head(string_counts_df, n =10)# Plot the top 10 most common strings using ggplot2ggplot(top_10_strings, aes(x =reorder(String, -Count), y = Count, fill = String)) +geom_bar(stat ="identity", position ="dodge", color ="black") +labs(title ="Top 10 Species hits",x = column_name,y ="Count") +theme_minimal() +theme(legend.position ="none") +coord_flip()``````{r go}data <-read.csv("../output/blast_annot_go.tab", sep ='\t')# Rename the `Gene.Ontology..biological.process.` column to `Biological_Process`colnames(data)[colnames(data) =="Gene.Ontology..biological.process."] <-"Biological_Process"# Separate the `Biological_Process` column into individual biological processesdata_separated <-unlist(strsplit(data$Biological_Process, split =";"))# Trim whitespace from the biological processesdata_separated <-gsub("^\\s+|\\s+$", "", data_separated)# Count the occurrences of each biological processprocess_counts <-table(data_separated)process_counts <-data.frame(Biological_Process =names(process_counts), Count =as.integer(process_counts))process_counts <- process_counts[order(-process_counts$Count), ]# Select the 20 most predominant biological processestop_20_processes <- process_counts[1:20, ]# Create a color palette for the barsbar_colors <-rainbow(nrow(top_20_processes))# Create a staggered vertical bar plot with different colors for each barbarplot(top_20_processes$Count, names.arg =rep("", nrow(top_20_processes)), col = bar_colors,ylim =c(0, max(top_20_processes$Count) *1.25),main ="Occurrences of the 20 Most Predominant Biological Processes", xlab ="Biological Process", ylab ="Count")# Create a separate plot for the legendpng("../output/GOlegend.png", width =800, height =600)par(mar =c(0, 0, 0, 0))plot.new()legend("center", legend = top_20_processes$Biological_Process, fill = bar_colors, cex =1, title ="Biological Processes")dev.off()``````{r legend, fig.width = 100 ,fig.height = 100}knitr::include_graphics("../output/GOlegend.png")```# Case Study - Oyster## Navigating AnnotationThe following is a stepwise example or annotation of a gene set using UniProt::Swiss-Prot (reviewed) such that Gene Ontology terms can be associated with each gene.In this following chunk where the fasta file is downloaded the [release](https://www.uniprot.org/help/release-statistics) is noted and the file name is modified accordingly.```{r engine='bash', eval=FALSE}cd DRAFT_Funct_Enrich/annotcurl -O https://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.fasta.gzmv uniprot_sprot.fasta.gz uniprot_sprot_r2023_02.fasta.gzgunzip -k uniprot_sprot_r2023_02.fasta.gz```A protein blast database is then made.```{r engine='bash', eval=FALSE}/home/shared/ncbi-blast-2.11.0+/bin/makeblastdb \-in DRAFT_Funct_Enrich/annot/uniprot_sprot_r2023_02.fasta \-dbtype prot \-out DRAFT_Funct_Enrich/annot/uniprot_sprot_r2023_02```In a majority of cases you will want to annotate a gene set to get gene ontology information. If you are creating your own genome or transcriptome it should be rather straightforward to know what file to annotate. If using a widely studied system where there are publically available resources, it is advisable to use those as this is the best way to facilitate integration of data sets. In this case study we will be considering the Eastern oyster, (*Crassostrea virginica*) for which there is data at [NCBI](https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_002022765.2/) and [Ensembl Metazoa](http://metazoa.ensembl.org/Crassostrea_virginica_gca002022765v4/Info/Index). At NCBI there is both a GenBank and RefSeq assembly available.In order to know which of the numerous fasta files should annotated with gene ontology information one should think downstream (or look to files already generated) to the identifiers in genesets that would be subject to functional enrichment tests.The resulting fpkm count matrix for our case study is from an experiment where male and female oysters where exposed to low pH (and control) conditions. The count matrix is accessible [here (csv)](https://github.com/epigeneticstoocean/2018_L18-adult-methylation/blob/main/analyses/gene_fpkm.csv). Hisat2/Stringtie was used to generate the count matrix with `GCF_002022765.2_C_virginica-3.0_genomic.gff` formatting thus responsible for gene naming. Specifically the naming format is as follows `gene-LOC111099033,gene-LOC111099034,gene-LOC111099035`.The following fasta was selected for annotation: `GCF_002022765.2_C_virginica-3.0_translated_cds.faa.gz````{r engine='bash', eval=FALSE}cd DRAFT_Funct_Enrich/annotcurl -O https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/022/765/GCF_002022765.2_C_virginica-3.0/GCF_002022765.2_C_virginica-3.0_translated_cds.faa.gzgunzip -k GCF_002022765.2_C_virginica-3.0_translated_cds.faa.gz``````{r engine='bash', eval=FALSE}head -2 DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.faaecho "number of sequences"grep -c ">" DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.faa`````` >lcl|NC_035780.1_prot_XP_022327646.1_1 [gene=LOC111126949] [db_xref=GeneID:111126949] [protein=UNC5C-like protein] [protein_id=XP_022327646.1] [location=join(30535..31557,31736..31887,31977..32565,32959..33204)] [gbkey=CDS]MTEVCYIWASSSTTVVICGIFFIVWRCFISIKKRASPLHGSSQQVCQTCQIEGHDFGEFQLSCRRQNTNVGYDLQGRRSD```This protein fasta is used as query for blast of uniprot_sprot database.```{r engine='bash', eval=FALSE}/home/shared/ncbi-blast-2.11.0+/bin/blastp \-query DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.faa \-db DRAFT_Funct_Enrich/annot/uniprot_sprot_r2023_02 \-out DRAFT_Funct_Enrich/annot/Cvir_transcds-uniprot_blastp.tab \-evalue 1E-20 \-num_threads 40 \-max_target_seqs 1 \-outfmt 6```Here is what the output file looks like, and at this point we want to get the UniProt Accession number for each gene```{r engine='bash', eval=FALSE}head -2 DRAFT_Funct_Enrich/annot/Cvir_transcds-uniprot_blastp.tab``````{r eval=FALSE}blast <-read.csv("DRAFT_Funct_Enrich/annot/Cvir_transcds-uniprot_blastp.tab", sep ='\t', header =FALSE)```Convert fasta to tab```{r engine='bash', eval=FALSE}perl -e '$count=0; $len=0; while(<>) {s/\r?\n//; s/\t/ /g; if (s/^>//) { if ($. != 1) {print "\n"} s/ |$/\t/; $count++; $_ .= "\t";} else {s/ //g; $len += length($_)} print $_;} print "\n"; warn "\nConverted $count FASTA records in $. lines to tabular format\nTotal sequence length: $len\n\n";' \DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.faa > DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.tab``````{r engine='bash', eval=FALSE}head -1 DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.tab``````{r}cdsftab <-read.csv("DRAFT_Funct_Enrich/annot/GCF_002022765.2_C_virginica-3.0_translated_cds.tab", sep ='\t', header =FALSE, row.names=NULL)```Now we can take the two data frames: A) blast output of taking protein fasta and comparing to uniprot_swiss-prot and B) a tabular version of same fasta file that has ID numbers of importance. Note this importance was determined based on what we want to use down stream.```{r, eval=FALSE}g.spid <-left_join(blast, cdsftab, by ="V1") %>%mutate(gene =str_extract(V2.y, "(?<=\\[gene=)\\w+")) %>%select(gene, V11, V2.x) %>%mutate(SPID =str_extract(V2.x, "(?<=\\|)[^\\|]*(?=\\|)")) %>%distinct(gene, SPID, .keep_all =TRUE)```Let's break it down step by step:1. **`g.spid <- left_join(blast, cdsftab, by = "V1")`** - This line is using the **`left_join()`** function from **`dplyr`** to merge the **`blast`** and **`cdsftab`** datasets by the column "V1". A left join retains all the rows in the **`blast`** data frame and appends the matching rows in the **`cdsftab`** data frame. If there is no match, the result is **`NA`**. The result of this operation is assigned to the **`g.spid`** object.2. **`mutate(gene = str_extract(V2.y, "(?<=\\[gene=)\\w+"))`** - This line is using the **`mutate()`** function from **`dplyr`** to add a new column called "gene" to the data frame. The new column is created by extracting substrings from the "V2.y" column based on the given regular expression pattern **`"(?<=\\[gene=)\\w+"`**. This regular expression matches and extracts any word (sequence of word characters, i.e., alphanumeric and underscore) that comes after "\[gene=".3. **`select(gene, V11, V2.x)`** - This line is using the **`select()`** function from **`dplyr`** to keep only the specified columns ("gene", "V11", and "V2.x") in the data frame.4. **`mutate(SPID = str_extract(V2.x, "(?<=\\|)[^\\|]*(?=\\|)"))`** - Again, the **`mutate()`** function is used to add another new column named "SPID". This column is created by extracting substrings from the "V2.x" column. The regular expression **`"(?<=\\|)[^\\|]*(?=\\|)"`** is designed to extract any character(s) that is/are surrounded by "\|" (pipe symbol). This is a common format for delimited strings.5. **`distinct(gene, SPID, .keep_all = TRUE)`** - This line is using the **`distinct()`** function from **`dplyr`** to remove duplicate rows based on the "gene" and "SPID" columns. The **`.keep_all = TRUE`** argument means that all other columns are also kept in the result, not just the "gene" and "SPID" columns.The resulting **`g.spid`** data frame should have unique rows with respect to the "gene" and "SPID" columns, and it should contain these two new columns, "gene" and "SPID", extracted from the original data based on specific string patterns.Now lets just write out SPIDs.```{r, eval=FALSE}left_join(blast, cdsftab, by ="V1") %>%mutate(gene =str_extract(V2.y, "(?<=\\[gene=)\\w+")) %>%select(gene, V11, V2.x) %>%mutate(SPID =str_extract(V2.x, "(?<=\\|)[^\\|]*(?=\\|)")) %>%distinct(gene, SPID, .keep_all =TRUE) %>%select(SPID) %>%write.table(file ="DRAFT_Funct_Enrich/annot/SPID.txt", sep ="\t", row.names =FALSE, quote =FALSE ) ```With a list of matching Swiss-Prot IDs, (technically UniProt Accession number) we can go back to https://www.uniprot.org and grab corresponding GO terms. This can be done via a web or using Python API.**Using Web**Using ID Mapping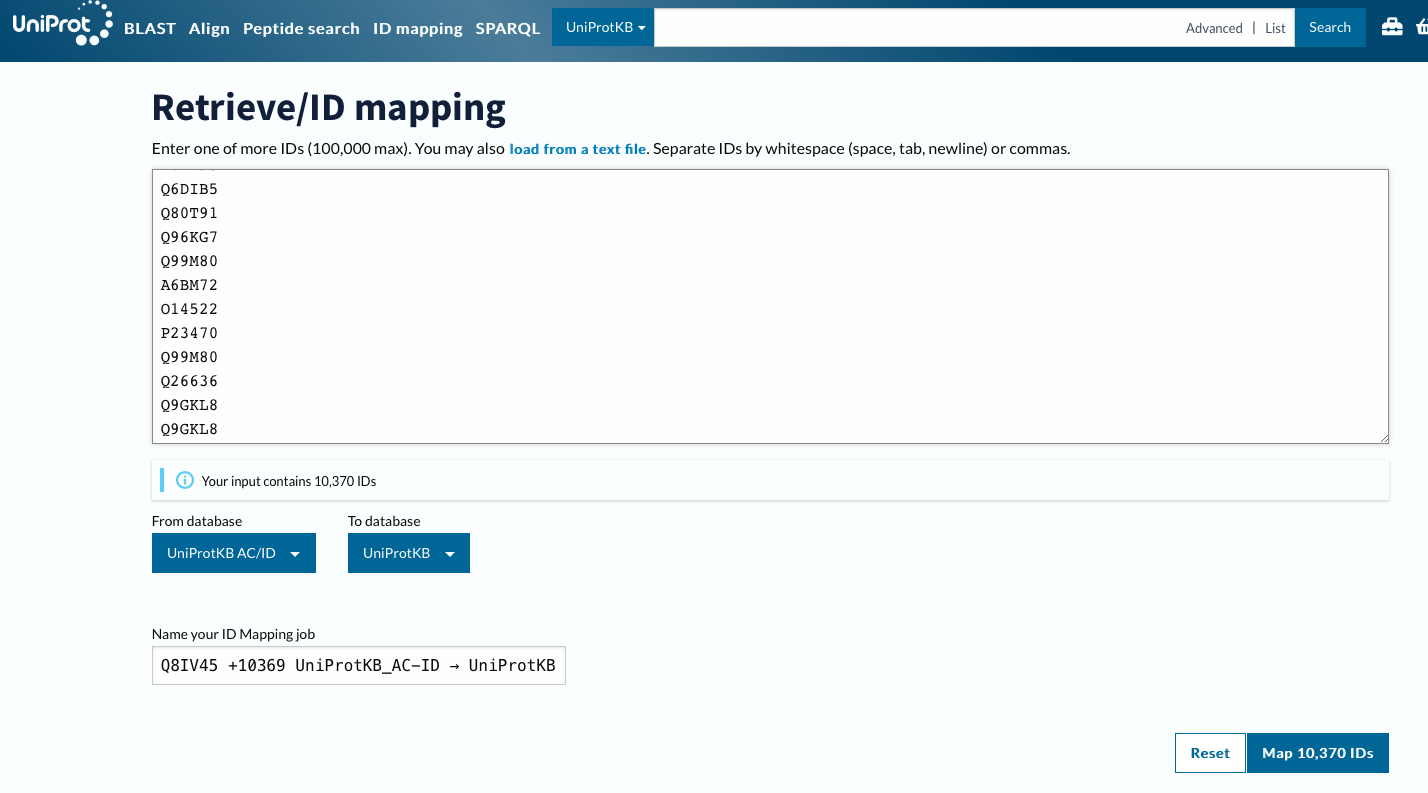Now will customize columns to get GO IDs.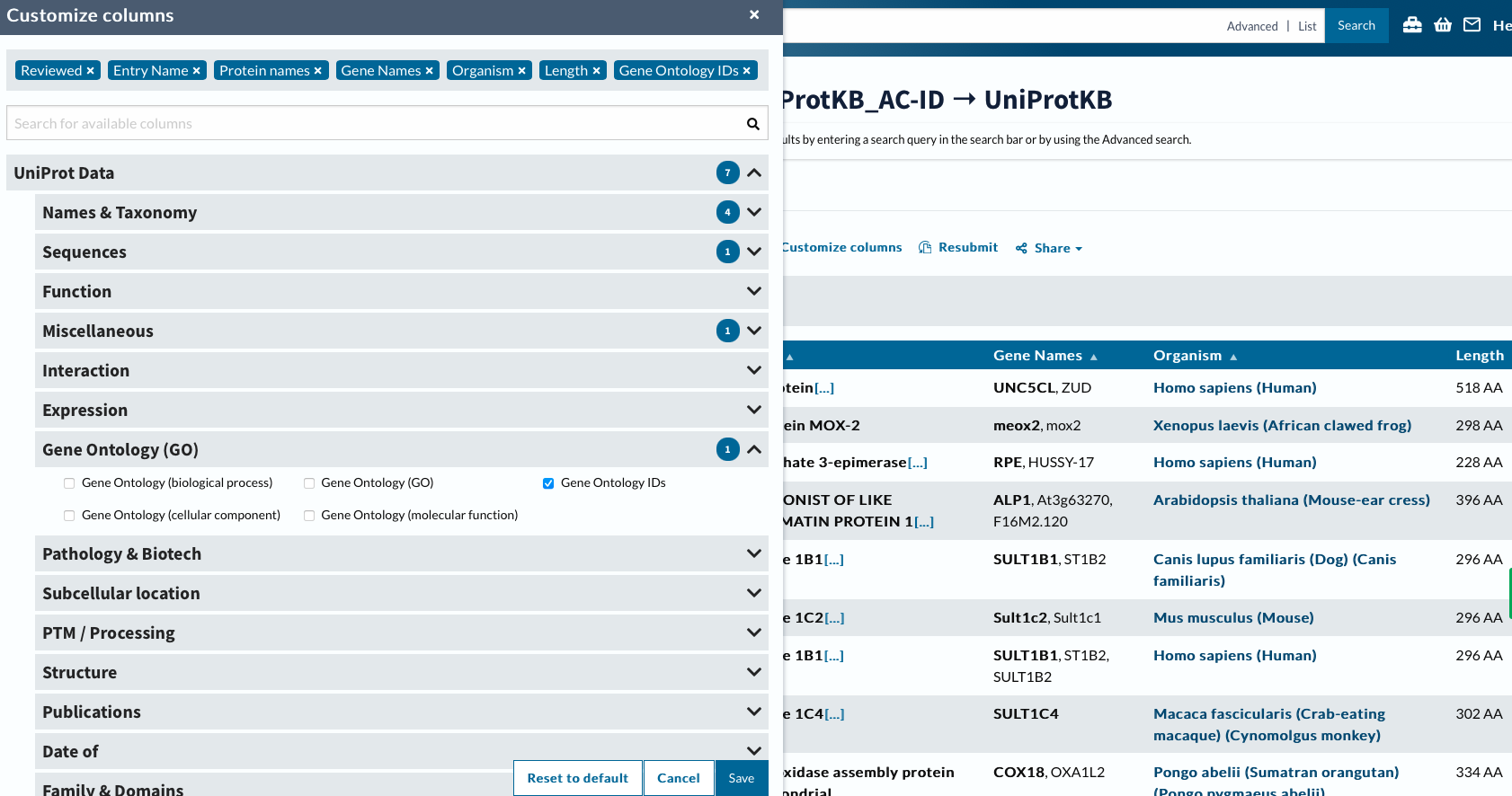```{r engine='bash', eval=FALSE}head -2 DRAFT_Funct_Enrich/annot/uniprotGO.tab```Finally we can join table to get "LOCIDs" the notation for our DEGs, with GO terms.```{r, eval=FALSE}go <-read.csv("DRAFT_Funct_Enrich/annot/uniprotGO.tab", sep ='\t', header =TRUE, row.names=NULL)``````{r, eval=FALSE}left_join(g.spid, go, by =c("SPID"="Entry")) %>%select(gene,Gene.Ontology.IDs) %>%write.table(file ="DRAFT_Funct_Enrich/annot/geneGO.txt", sep ="\t", row.names =FALSE, quote =FALSE )``````{r engine='bash', eval=FALSE}head DRAFT_Funct_Enrich/annot/geneGO.txt```**Using API**```{r engine='bash'}python3 DRAFT_Funct_Enrich/annot/uniprot-retrieval.py DRAFT_Funct_Enrich/annot/SPID.txt```