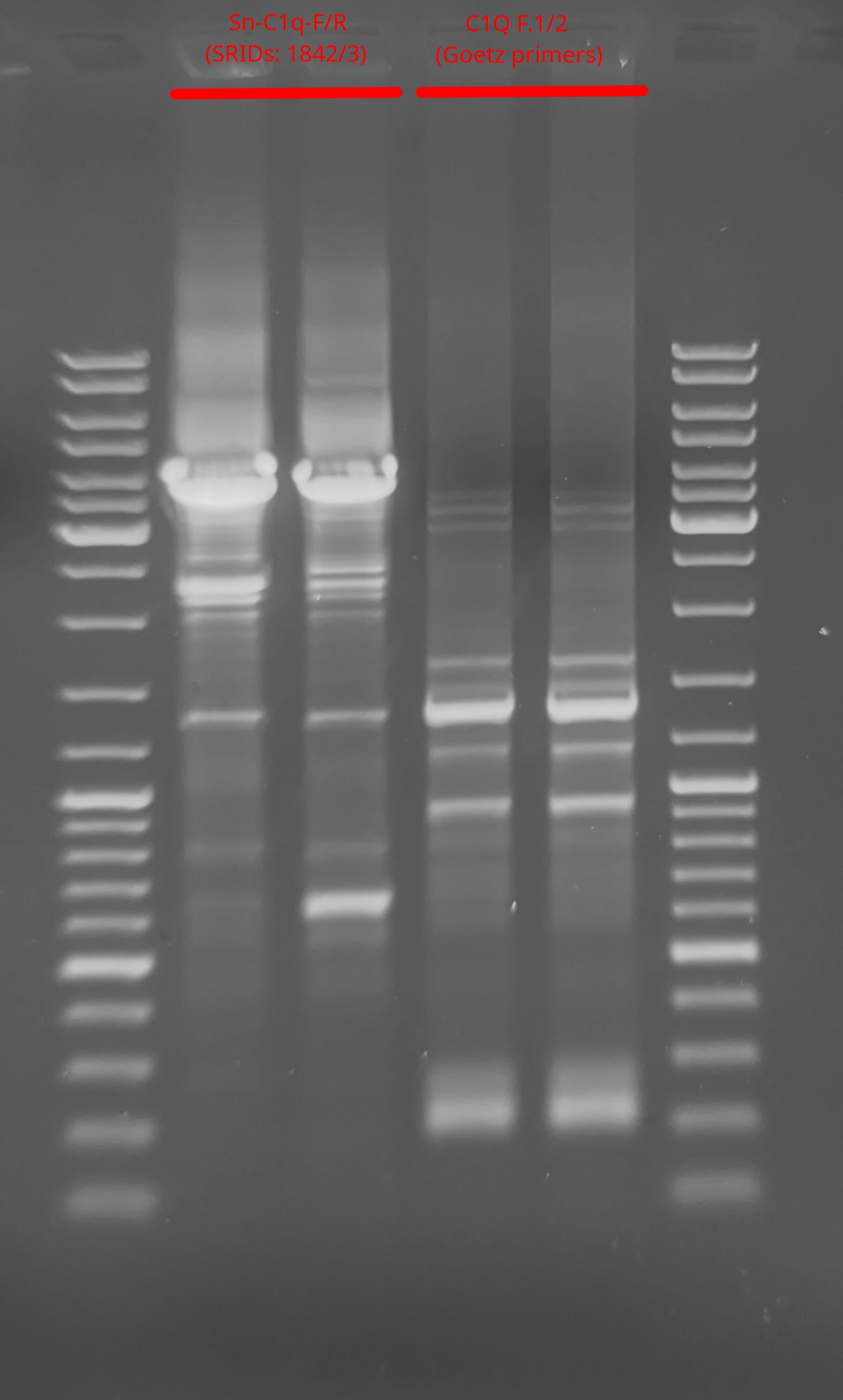
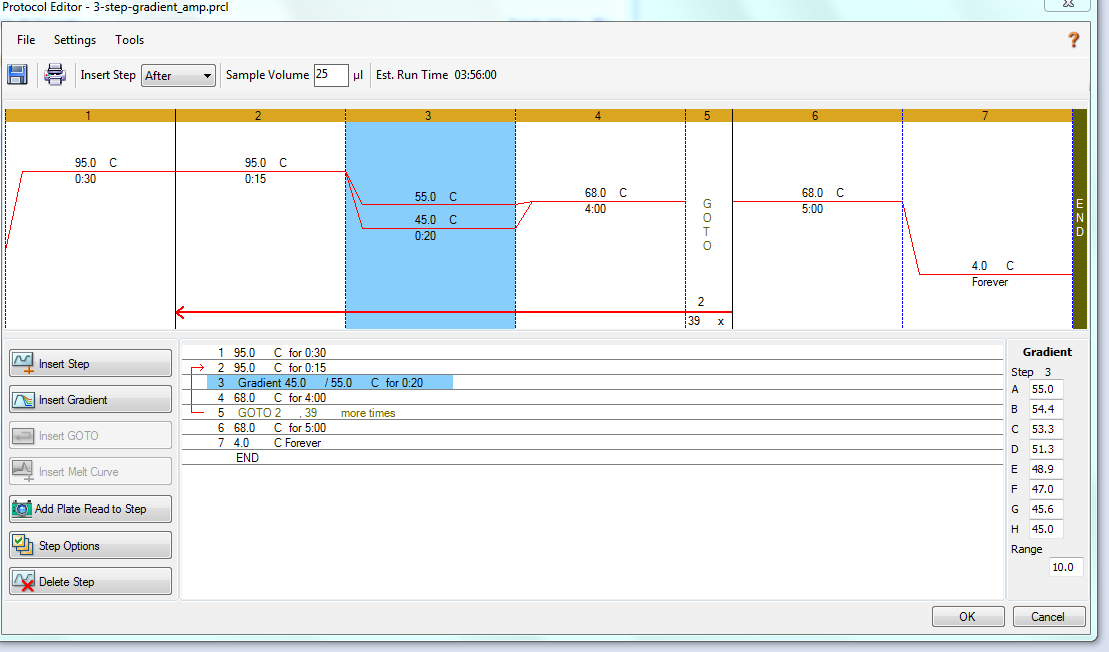
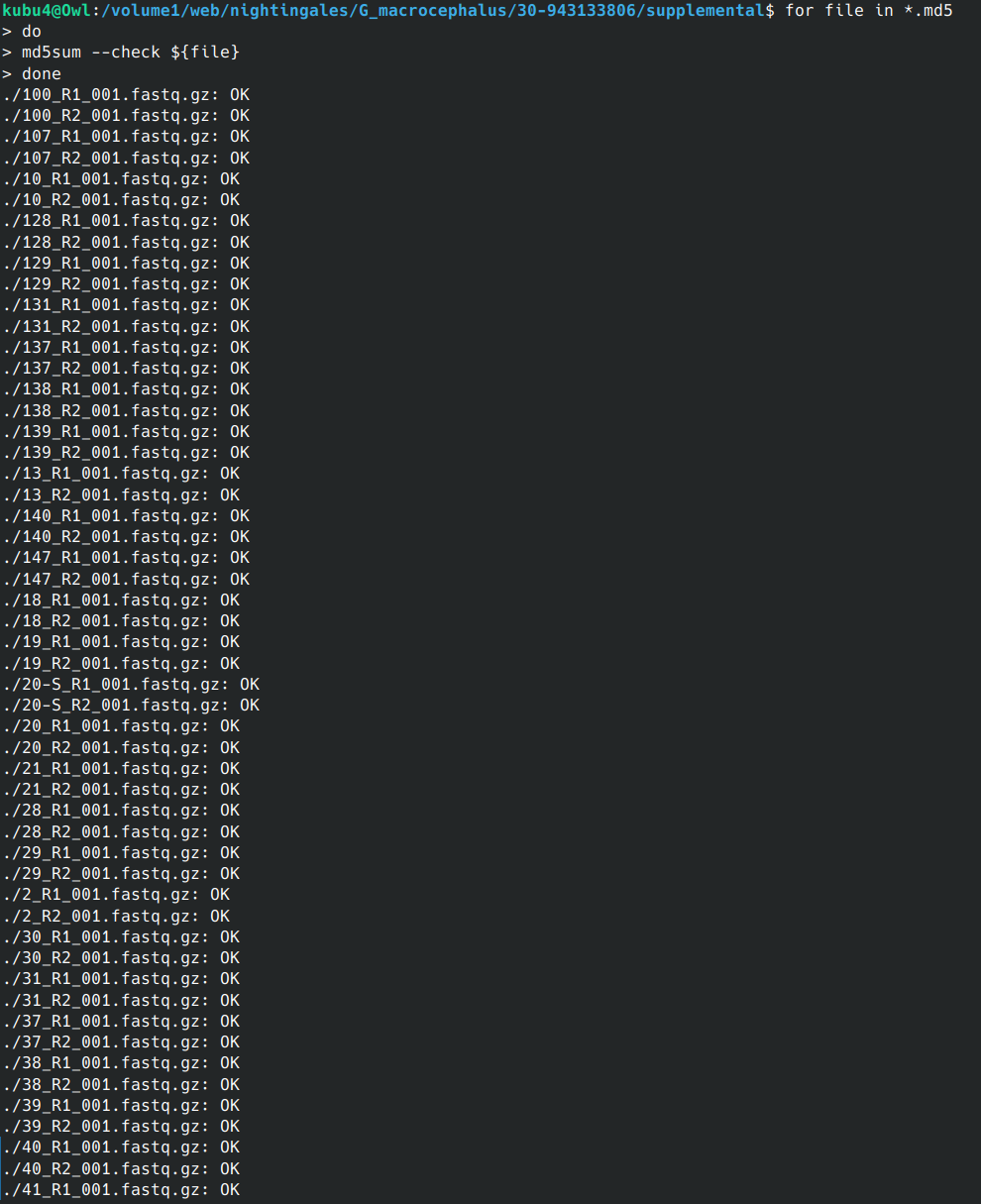
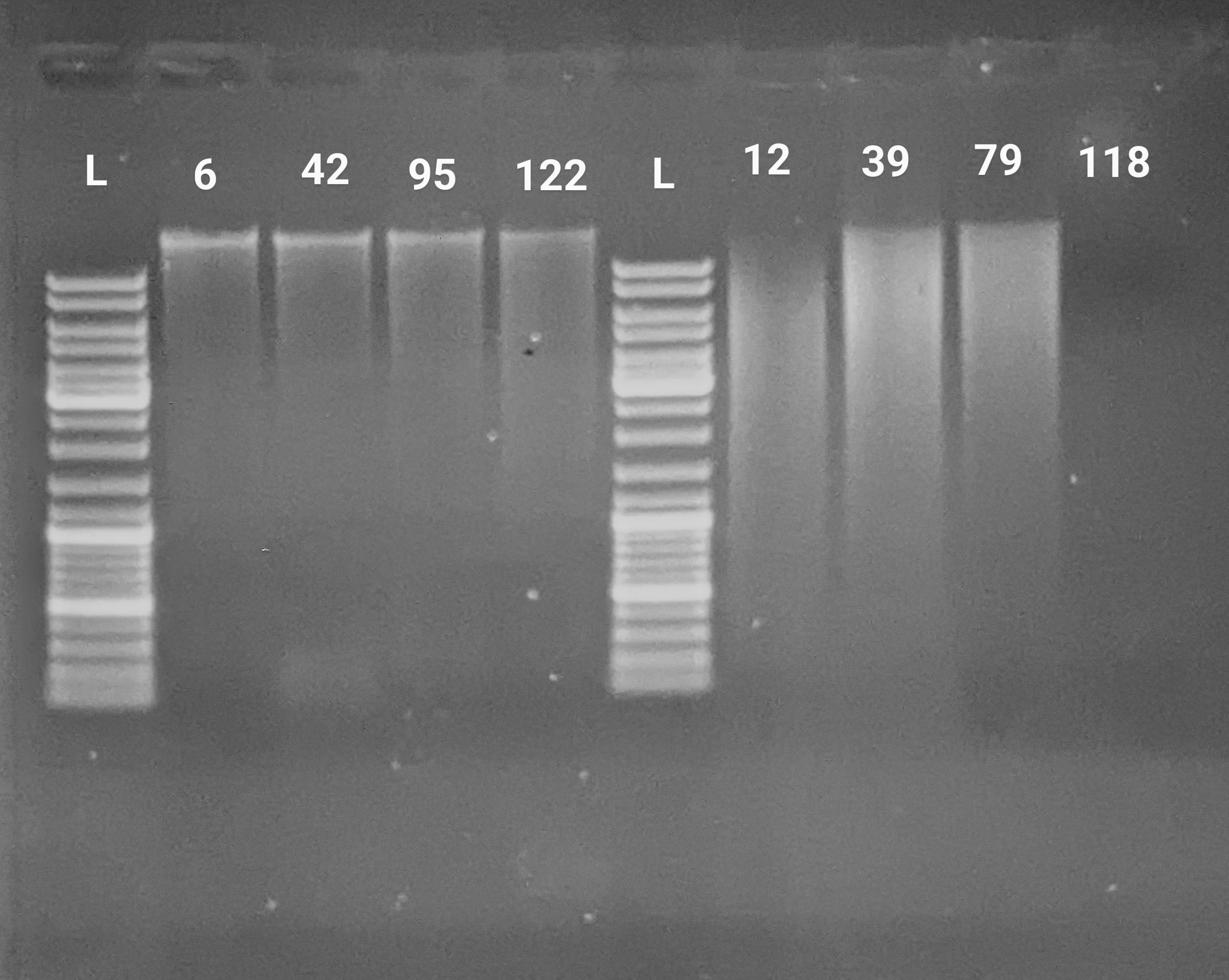
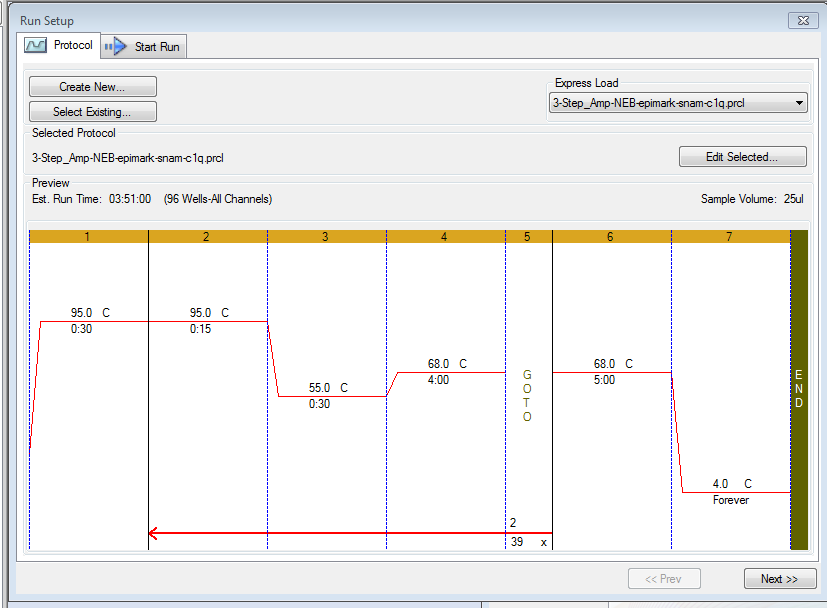
PCR - Lake Trout C1q Primers with Bisulfite-treated gDNA
2024
C1q
Lake trout
Salvelinus namaycush
PCR
bisulfite
gDNA
liver
gel
RNA-seq Alignment - A.pulchra RNA-seq Alignments Using HISAT2 and StringTie for Azenta Project 30-1047560508
2024
30-1047560508
RNA-seq
E5
timeseries_molecular
HISAT2
StringTie
alignment
Acropora pulchra
coral
Data Wrangling - Generate GTF from A.pulchra Genome GFF Using gffread
2024
coral
Acropora pulchra
genome
GFF
GTF
gffread
E5
timeseries_molecular
Genome Indexing - A.pulchra Genome Using HISAT2
2024
coral
Acropora pulchra
HISAT2
E5
timeseries_molecular
FastQC Trimming and QC - A.pulchra RNA-seq from Azenta Project 30-1047560508 Using fastp
2024
RNA-seq
30-1047560508
Azenta
coral
Acropora pulchra
fastp
FastQC
MultiQC
trimming
E5
timeseries_molecular
FastQ QC - A.pulchra RNA-seq from Azenta Project 30-1047560508
2024
RNA-seq
30-1047560508
Azenta
coral
Acropora pulchra
FastQC
MultiQC
timeseries_molecular
E5
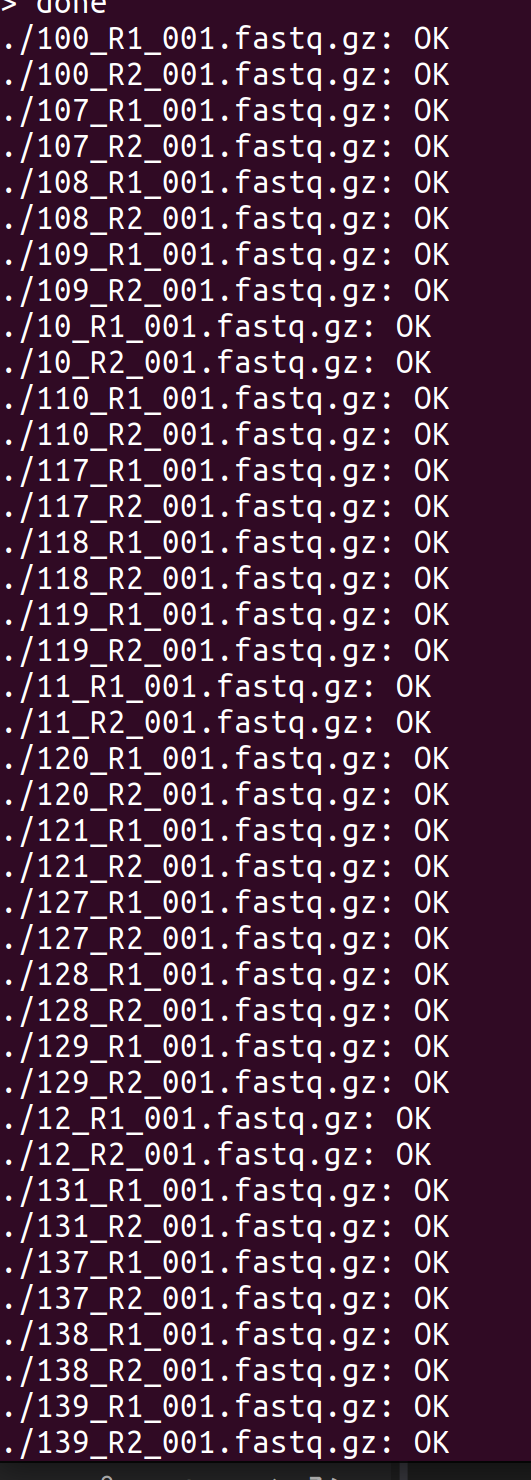
Data Received - R.philippinarum (Manilla clam) sRNA-seq Data from Azenta Project 30-1035633055
2024
Data Received
sRNA-seq
30-1035633055
Ruditapes philippinarum
Manila clam
Agarose Gel - gDNA Integrity Check of Lake Trout Liver gDNA from 20240712
2024
agarose gel
gDNA
Lake trout
liver
Salvelinus namaycush
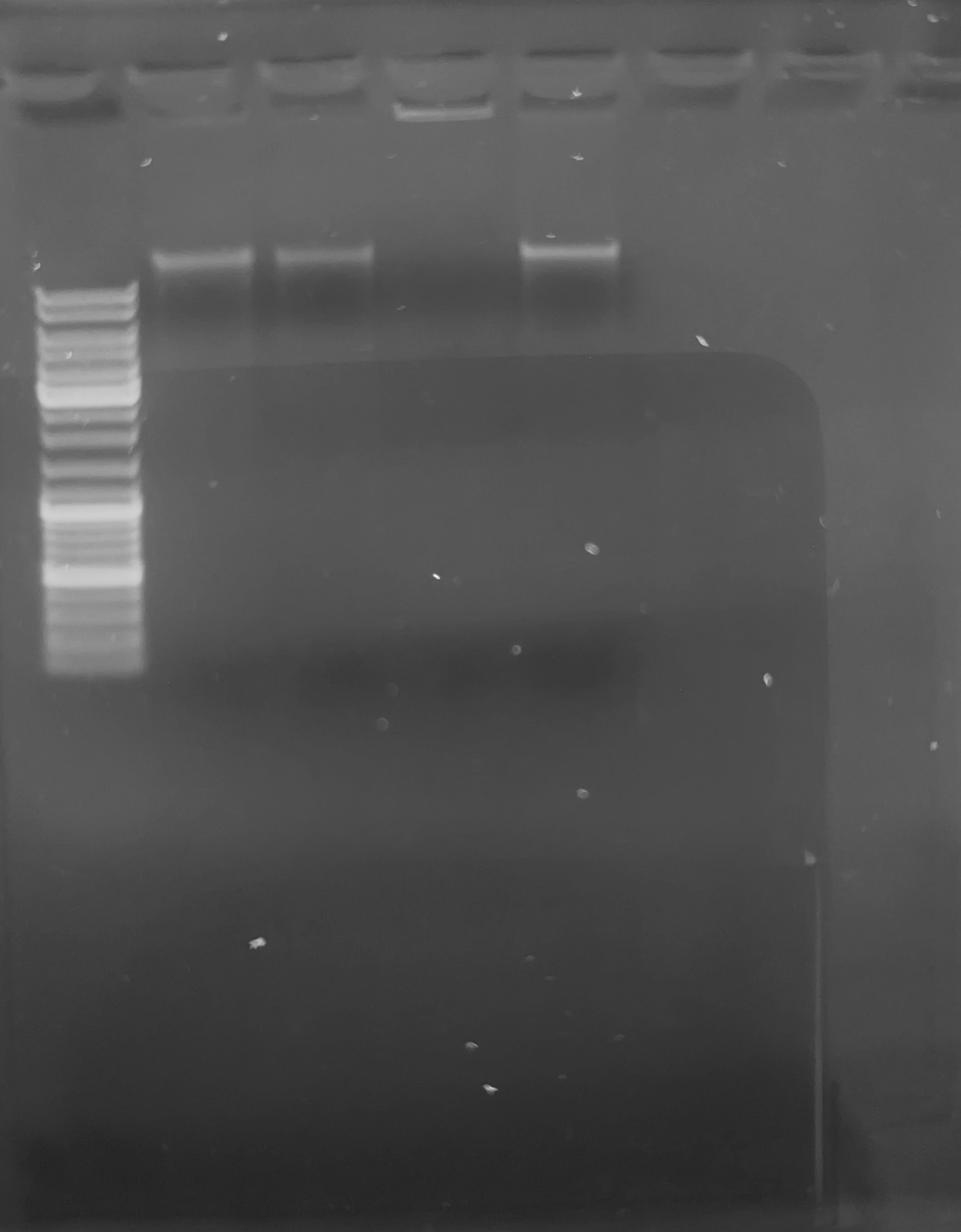
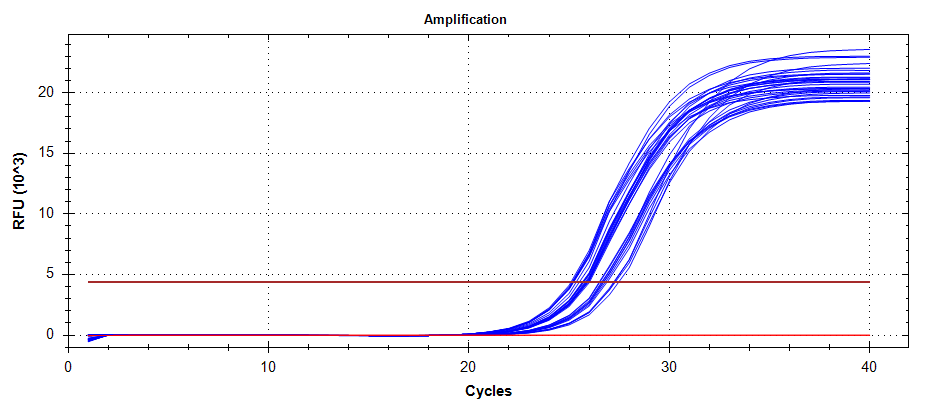
PCR - Lake Trout C1Q with Liver gDNA
2024
PCR
Lake trout
C1q
CFX Connect
EpiMark Hot Start Taq
gDNA
liver
Salvelinus namaycush
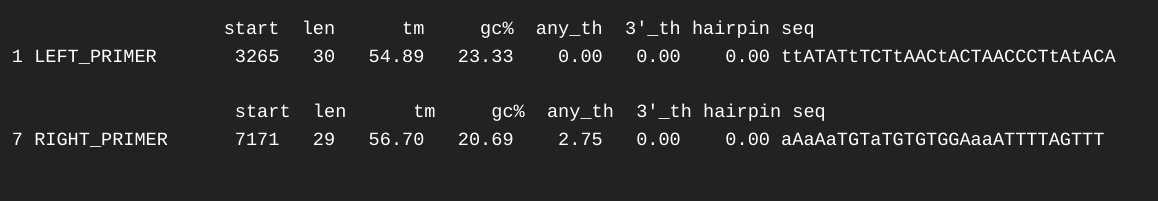
Primer Design - Lake Trout C1q Gene Sequencing Primers Using Primer3
2024
C1q
lake trout
Salvelinus namaycush
Primer3
primer design
EMBOSS
pyfaidx
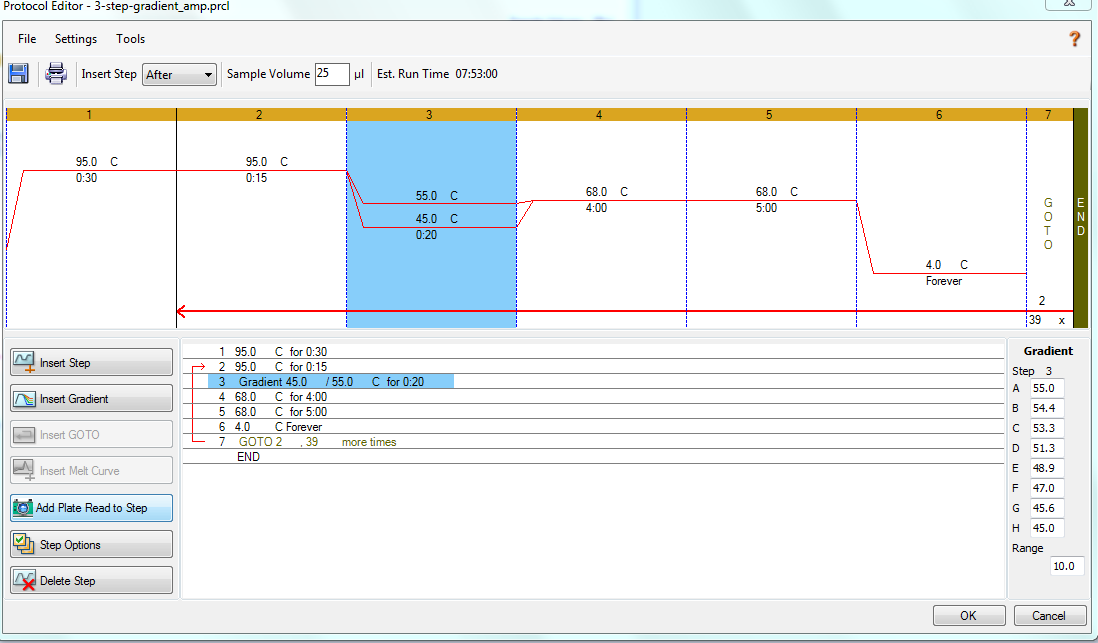
Bisulfite PCR - Second Primer Annealing Gradient Test with Lake Trout Bisulfite-treated DNA
2024
bisulfite
BS-PCR
PCR
DNA
Lake troute
Salvelinus namaycush
Data Received - G.macrocephalus Supplemental RNA-seq Data from Azenta Project 30-943133806
Data Received
2024
Gadus macrocephalus
Pacific cod
RNA-seq
Azenta
DNA and RNA Isolations - Gadus macrocephalus Blood
2024
DNA isolation
RNA isolation
Gadus macrocephalus
Pacific cod
blood
Zymo Quick-DNA/RNA Miniprep
Qubit 3.0
Qubit DNA BR Assay
Qubit RNA HS Assay
Bisulfite PCR - Testing Primer Annealing Gradient with Lake Trout Bisulfite-treated DNA
2024
BS-PCR
bisulfite
PCR
DNA
Lake trout
Salvelinus namaycush
Bisulfite Conversion - Lake Trout gDNA Using EZ DNA Methylation Lightning Kit
2024
lake trout
Salvelinus namaycush
bisulfite
EZ DNA Methylation Lightning Kit
Primer Design - Lake Trout C1q Gene Bisulfite Sequencing Primers Using Primer3
2024
C1q
lake trout
Salvelinus namaycush
Primer3
primer design
Samples Submitted - Macs Manilla Clam Egg Samples for sRNA-seq at Azenta
2024
Samples Submitted
Azenta
sRNA-seq
Manila clam
Lajonkairia lajonkairii
Ruditapes philippinarum
eggs
DNA Isolation Quantification and Integrity Assessment - Pacific Cod Blood Samples Test
2024
Pacific cod
Gadus macrocephalus
DNA isolation
DNA quantification
Qubit
DNA BR Assay
agarose gel
DNA Isolation and Quantification - Pacific Cod Blood
2024
Pacific cod
Gadus macrocephalus
blood
DNA isolation
DNA quantification
Qubit
DNA BR assay
DNA Isolation and Quantification - Lake Trout Liver
DNA isolation
2024
lake trout
Salvelinus namaycush
Qubit
DNA BR assay
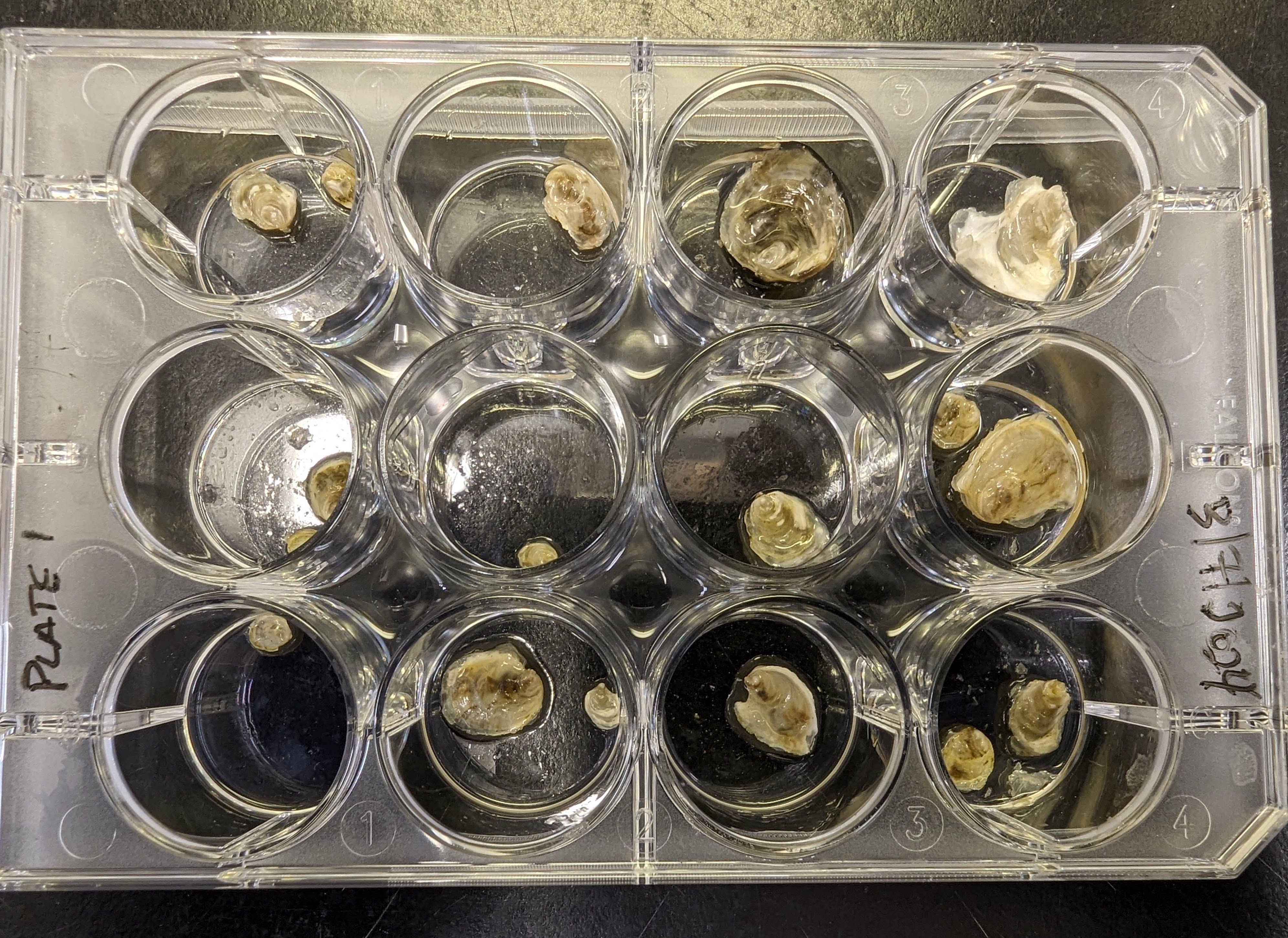
Samples Received - L.staminea Histology Slides for Mac
2024
Leukoma staminea
littleneck clam
histology
slides
Samples Received
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-lifestage-carryover
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
RNA HS assay
Qubit
RNA isolation
RNA quantification
Directzol RNA miniprep
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-lifestage-carryover
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
RNA HS assay
Qubit
RNA isolation
Directzol RNA Miniprep
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-lifestage-carryover
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
RNA HS assay
Qubit
RNA isolation
Directzol RNA Miniprep
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-lifestage-carryover
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
RNA HS assay
Qubit
RNA isolation
Directzol RNA Miniprep
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-lifestage-carryover
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
RNA HS assay
Qubit
RNA isolation
Directzol RNA Miniprep
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-gigas-carryover
Crassostrea gigas
Pacific oyster
adult
seed
spat
Directzol RNA Miniprep
Qubit
RNA HS Assay
RNA Isolation and Quantification - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-gigas-carryover
Crassostrea gigas
Pacific oyster
adult
seed
spat
Directzol RNA Miniprep
Qubit
RNA HS assay
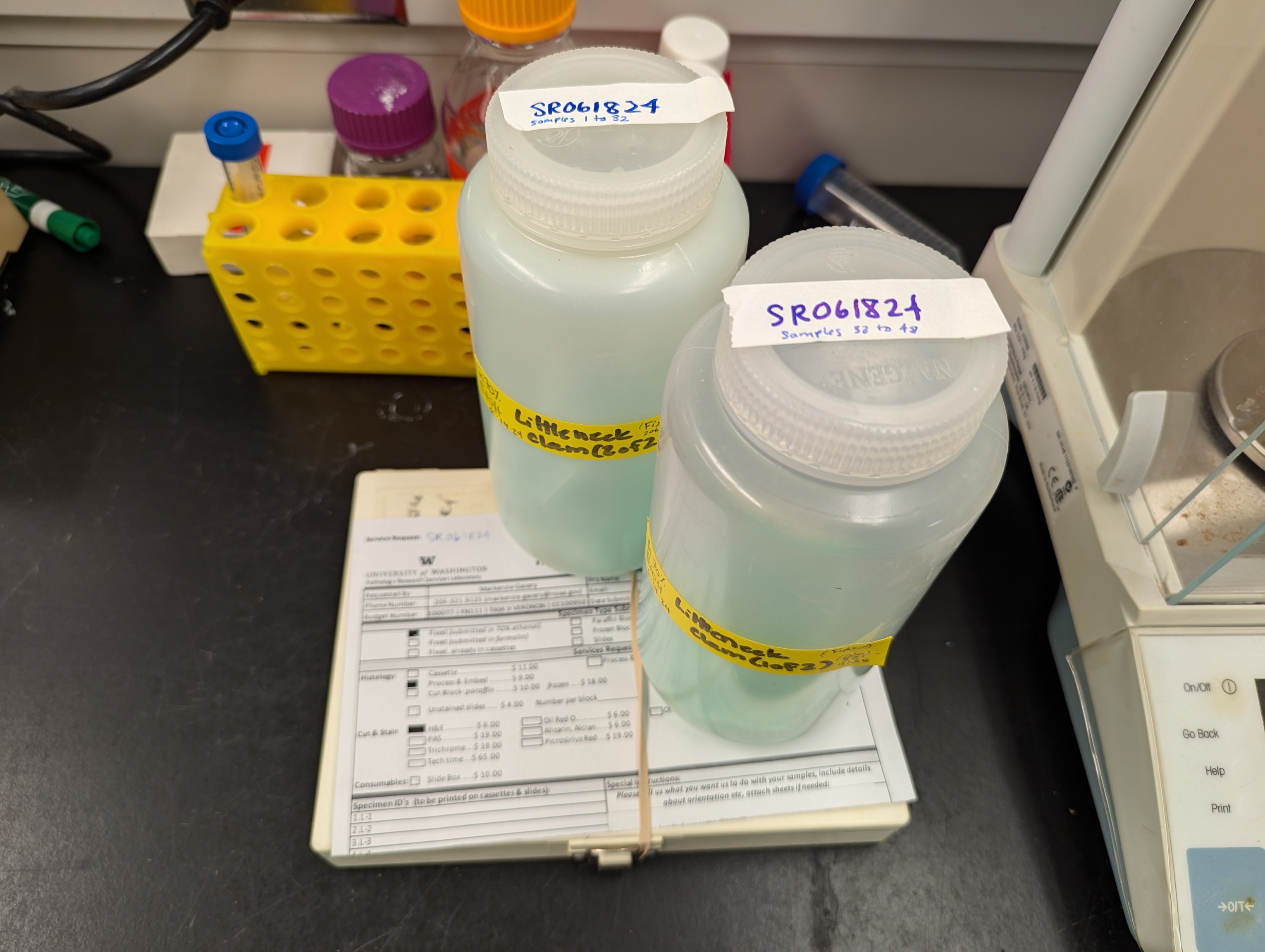
Samples Submitted - Littleneck Clam Histology Cassettes to UW Pathology Research Services Laboratory
2024
histology
littleneck clam
Pathology Research Services Laboratory
Samples Submitted
RNA Quantification - C.gigas RNA Lifestage Carryover from 20240615 and 20240617
2024
project-gigas-carryover
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
Qubit
RNA HS assay
RNA Isolation - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-gigas-carryover
Directzol RNA Miniprep
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
RNA Isolation - C.gigas Lifestage Carryover Seed Juvenile and Adult
2024
project-gigas-carryover
Directzol RNA Miniprep
Crassostrea gigas
Pacific oyster
seed
adult
juvenile
Differential Gene Expression - G.macrocephalus RNA-seq 9C vs 16C Using edgeR
edgeR
Pacific cod
RNA-seq
differential gene expression
2024
Gadus macrocephalus
Samples Received - Lajonkairia lajonkairii Egg Samples from Mackenzie Gavery
Lajonkairia lajonkairii
manilla clam
2024
Samples Received
Agarose Gel - G.macrocephalus gDNA from 20240521
agarose
gDNA
Gadus macrocephalus
Pacific cod
gel
2024
DNA Isolation - Pacific cod liver
DNA isolation
Pacific cod
liver
Gadus macrocephalus
DNA quantification
Qubit
2024
Samples Submitted - Pacific Cod 40 Tissue Samples for WGBS at Psomagen
psomagen
WGBS
2024
Pacific cod
Gadus macrocephalus
BS-seq
bisulfite
Samples Submitted
qPCR Analysis - C.gigas Lifestages Carryover from 20240325
2024
qPCR
Crassotrea gigas
Pacific oyster
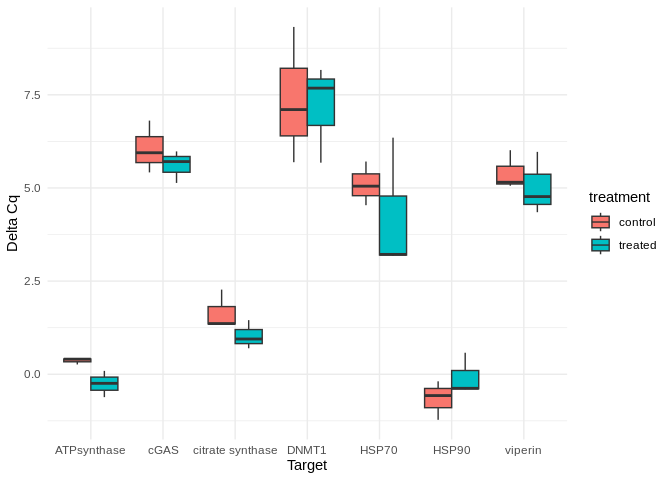
qPCRs - C.gigas Lifestage Carryover cDNA
qPCR
SsoFast
CFX Connect
HSP70
HSP90
GAPDH
VIPERIN
ATPsynthase
cGAS
DMNT1
citrate synthase
Crassostrea gigas
Pacific oyster
cDNA
Reverse Transcription - C.gigas Lifestage Carryover Seed and Spat
reverse transcription
Lifestage carryover
Crassostrea gigas
Pacific oyster
cDNA
M-MLV
2024
RNA Isolation - C.gigas Lifestage Carryover Seed and Spat
RNA isolation
Crassostrea gigas
Lifestage carryover
Pacific oyster
2024
SRA Submission - CEABIGR WGBS and RNA-seq
SRA
CEABIGR
Crassostrea virginica
Eastern oyster
RNAseq
WGBS
2024
Data Received - Pacific cod RNA-seq Azenta Project 30-943133806
Azenta
RNA-seq
2024
Pacific cod
Gadus macrocephalus
30-943133806
Data Received
Figure Updates - CEABIGR Spurious Transcription Calcs and Plotting Using Exon Sum Threshold
CEABIGR
2024
plot
Eastern oyster
Crassostrea virginica
Data Exploration - CEABIGR Spurious Transcription Calculations and Plotting
2024
CEABIGR
plot
Eastern oyster
Crassostrea virginica
Samples Submitted - Gadus macrocephalus liver tissues to Azenta for RNA-seq
2023
Gadus macrocephalus
Pacific cod
Azenta
RNA-seq
Samples Submitted
liver
spleen
gill
blood
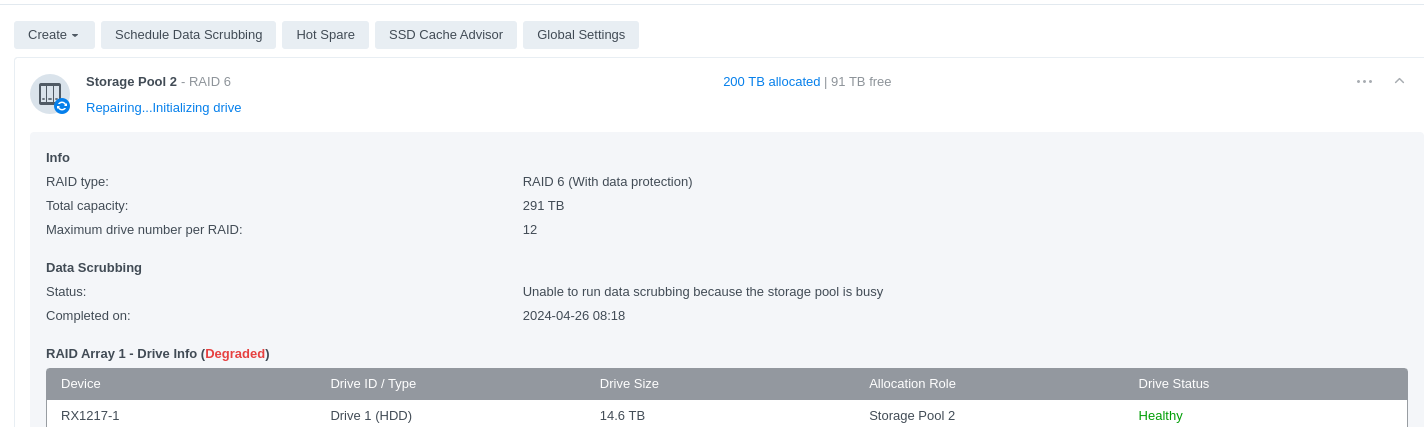
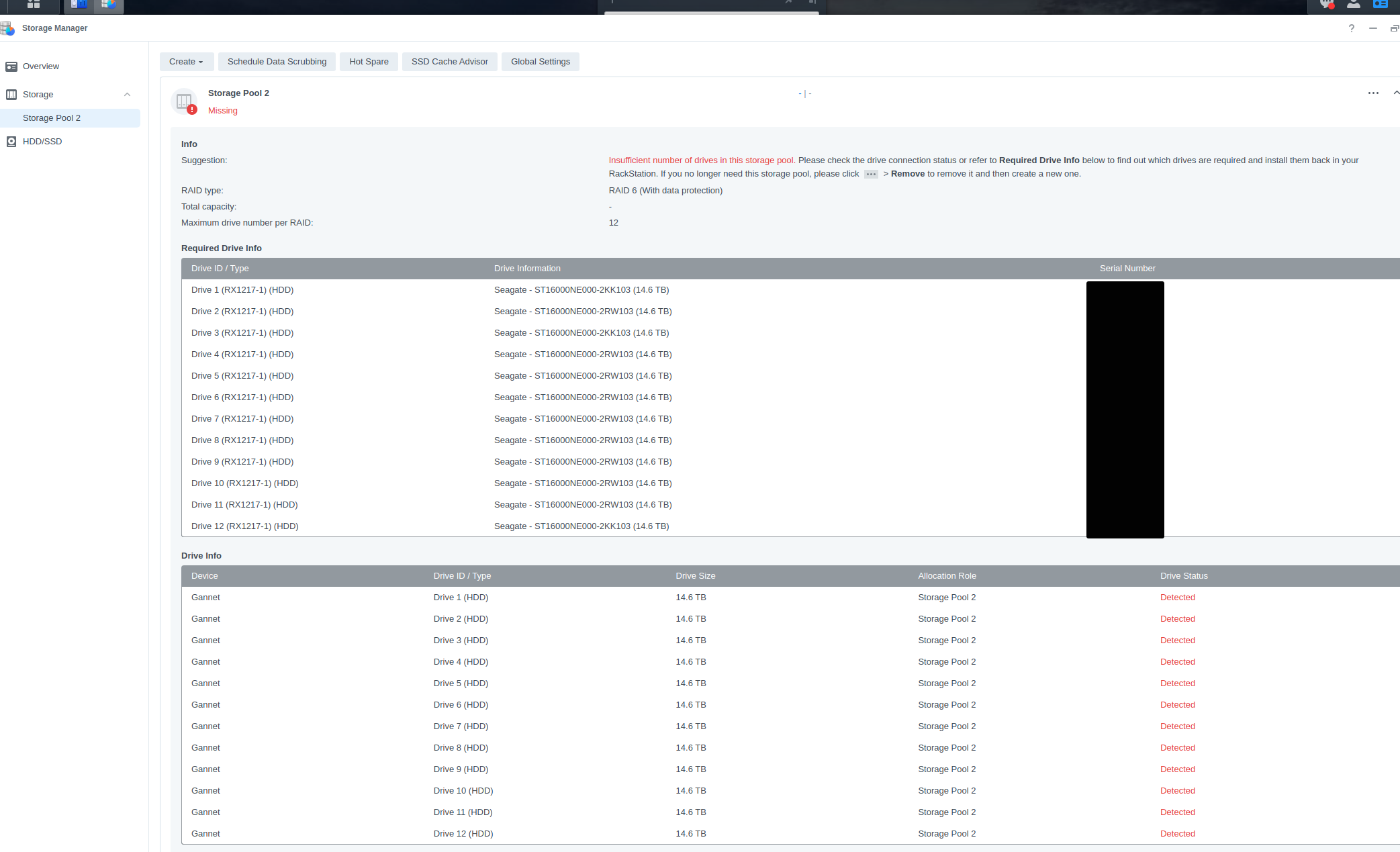
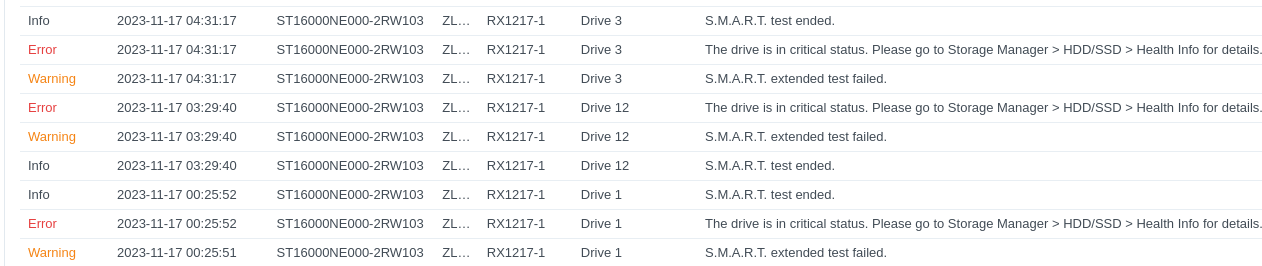
Computer Maintenance - Replace Failing HDDs in Synology RX1217 Expansion Unit on Gannet
gannet
synology
RX1217
HDDs
2023
computer maintenance
Transcriptome Annotation - M.magister De Novo Transcriptome Assembly Using Trinotate on Mox
mox
2023
Metacarcinus magister
Dungenss crab
Trinotate
transcriptome
annotation
Transcriptome Annotation - M.magister De Novo Transcriptome Assembly for DuMOAR Project Using DIAMOND BLASTx on Mox
mox
DIAMOND
BLASTx
transcriptome
2023
DuMOAR
Dungeness crab
Metacarcinus magister
Assembly Comparisons - Ostrea lurida Non-scaffold Genome Assembly Comparisons Using Quast on Swoose
2021
Olympia Oyster Genome Assembly
Genome Submission - Validation of Olurida_v081.fa and Annotated GFFs Prior to Submission to NCBI
2021
Olympia Oyster Genome Assembly
ENA Submission - Ostrea lurida draft genome Olurida_v081.fa
2020
Olympia Oyster Genome Sequencing
Data Received - C.bairdi RNAseq Data from Genewiz
2020
Data Received
Tanner Crab RNAseq
Data Wrangling - Renaming, Splitting, and Feature Counts of Updated Pgenerosa_v074 GenSAS Merged GFF
2019
Geoduck Genome Sequencing
Data Received - C.bairdi RNAseq Day9-12-26 Infected-Uninfected
2019
Tanner Crab RNAseq
Data Received
Genome Annotation - O.lurida 20190709-v081 Transcript Isoform ID with Stringtie on Mox
2019
Olympia Oyster Genome Sequencing
Sample Submission - Tanner Crab Infected vs Uninfected RNAseq
2019
Tanner Crab RNAseq
Samples Submitted
SRA Submission - Olymia oyster Whole Genome BS-seq Data
2018
Olympia oyster reciprocal transplant
TrimGalore/FastQC/MultiQC - C.virginica Oil Spill MBDseq Concatenated Sequences
2018
LSU C.virginica Oil Spill MBD BS Sequencing
Genome Annotation – Olympia oyster genome annotation results #02
2018
Olympia Oyster Genome Sequencing
Genome Annotation - Olympia oyster genome annotation results #01
2018
Olympia Oyster Genome Sequencing
Mox – Over quota: Olympia oyster genome annotation progress (using Maker 2.31.10)
2018
Olympia Oyster Genome Sequencing
BS-seq Mapping - Olympia oyster bisulfite sequencing: TrimGalore > FastQC > Bismark
2018
BS-seq Libraries for Sequencing at Genewiz
MBD Enrichment for Sequencing at ZymoResearch
Data Management - Geoduck Phase Genomics Hi-C Data
2018
Geoduck Genome Sequencing
Samples Received
Data Management - SRA Submission LSU C.virginica Oil Spill MBD BS-seq Data
2018
LSU C.virginica Oil Spill MBD BS Sequencing
Genome Assembly - SparseAssembler Geoduck Genomic Data, kmer=101
2018
Geoduck Genome Sequencing
Assembly Comparisons – Oly Assemblies Using Quast
2018
Olympia Oyster Genome Sequencing
Samples Submitted - Pulverized Geoduck Tissues to Illumina for More 10x Genomics Sequencing
2017
Geoduck Genome Sequencing
Samples Submitted
Genome Assembly – Olympia Oyster Illumina & PacBio Using PB Jelly w/BGI Scaffold Assembly
2017
Olympia Oyster Genome Sequencing
Samples Submitted - Geoduck Tissues to Illumina for More 10x Genomics Sequencing
2017
Geoduck Genome Sequencing
Samples Submitted
Assembly Comparison - Oly Assemblies Using Quast
2017
Olympia Oyster Genome Sequencing
Genome Assembly - Olympia oyster Illumina & PacBio reads using MaSuRCA
2017
Olympia Oyster Genome Sequencing
Genome Assembly - Olympia Oyster Redundans with Illumina + PacBio
2017
Olympia Oyster Genome Sequencing
Genome Assembly - minimap/miniasm/racon Overview
2017
Olympia Oyster Genome Sequencing
Samples Submitted - Geoduck Ctenidia to Illumina for 10x Genomics Sequencing
2017
Geoduck Genome Sequencing
Samples Submitted
Project Progress - Olympia Oyster Genome Assemblies by Sean Bennett
2017
Olympia Oyster Genome Sequencing
Manuscript Writing - Submitted!
2017
Genotype-by-sequencing at BGI
Manuscript - Oly GBS 14 Day Plan
2017
Genotype-by-sequencing at BGI
Data Management – SRA Submission Oly GBS Batch Submission
2017
Genotype-by-sequencing at BGI
Sample Submission – Geoduck gDNA for Illumina Pilot Sequencing Project
2017
Geoduck Genome Sequencing
Samples Submitted
Sample Submission - Geoduck Tissue & gDNA for Illumina Pilot Sequencing Project
2016
Geoduck Genome Sequencing
Samples Submitted
Sample Submission - Ostrea lurida gDNA for PacBio Sequencing
2016
Olympia Oyster Genome Sequencing
Samples Submitted
Data Management - Integrity Check of Final BGI Olympia Oyster & Geoduck Data
2016
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Data Management - Download Final BGI Genome & Assembly Files
2016
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Data Analysis - Continued O.lurida Fst Analysis from GBS Data
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Data Analysis - Initial O.lurida Fst Determination from GBS Data
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Data Management - Tracking O.lurida FASTQ File Corruption
2016
Olympia Oyster Genome Sequencing
Computing - Retrieve data from Amazon EC2 Instance
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Server HDD Failure – Owl
2016
Computer Servicing
Computing - Amazon EC2 Instance Out of Space?
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
SRA Release - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck)
2016
Protein expression profiles during sexual maturation in Geoduck
SRA Submissions
Data Management - O.lurida Raw BGI GBS FASTQ Data
2016
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Data Management - Concatenate FASTQ files from Oly MBDseq Project
2016
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
SRA Submission – Genome sequencing of the Olympia oyster (Ostrea lurida)
2016
Olympia Oyster Genome Sequencing
SRA Submissions
SRA Submission - Transcriptomic Profiles of Adult Female & Male Gonads in Panopea generosa (Pacific geoduck).
2016
Protein expression profiles during sexual maturation in Geoduck
Data Management - O.lurida 2bRAD Dec2015 Undetermined FASTQ files
2016
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Data Received - Ostrea lurida MBD-enriched BS-seq
2016
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Data Received - Bisulfite-treated Illumina Sequencing from Genewiz
2016
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Data Received - Oly 2bRAD Illumina Sequencing from Genewiz
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Sample Submission - BS-seq Library Pool to Genewiz
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Samples Submitted
Illumina Methylation Library Quantification - BS-seq Oly/C.gigas Libraries
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Illumina Methylation Library Construction - Oly/C.gigas Bisulfite-treated DNA
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Bioanalyzer - Bisulfite-treated Oly/C.gigas DNA
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Bisulfite Treatment - Oly Reciprocal Transplant DNA & C.gigas Lotterhos DNA for BS-seq
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Agarose Gel - Oly gDNA for BS-seq Libraries, Take Two
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Agarose Gel - Oly gDNA for BS-seq Libraries
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
DNA Isolation - Oly gDNA for BS-seq
2015
BS-seq Libraries for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Sample Submission - 2bRAD Libraries for Genewiz
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Samples Submitted
Sample Submission - Olympia oyster MBD-enriched DNA to ZymoResearch
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Samples Submitted
Sample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI
2015
Olympia Oyster Genome Sequencing
Samples Submitted
Sample Submission - Oly Oyster Bay Tissues for GBS
2015
Genotype-by-sequencing at BGI
Olympia oyster reciprocal transplant
Samples Submitted
Samples Received - Oly Tissue & DNA from Katherine Silliman
2015
Olympia oyster reciprocal transplant
Samples Received
DNA Quality Assessment - Geoduck & Olympia Oyster gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
DNA Quantification - MBD-enriched Olympia oyster DNA
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
Ethanol Precipitation - Olympia oyster MBD
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
MBD Enrichment - Sonicated Olympia Oyster gDNA
2015
MBD Enrichment for Sequencing at ZymoResearch
Olympia oyster reciprocal transplant
DNA Sonication - Oly gDNA for MBD
2015
MBD Enrichment for Sequencing at ZymoResearch
qPCR – Oly RAD-Seq Library Quantification
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
qPCR - Oly RAD-Seq Library Quantification
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Gel Extraction - Oly RAD-Seq Prep Scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
DNA Quality Assessment - Geoduck, Oly & Oly 2SN
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Olympia oyster reciprocal transplant
PCR – Oly RAD-seq Prep Scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
PCR – Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
DNA Quantification & Quality Assessment - Oly 2SN gDNA
2015
Olympia oyster reciprocal transplant
DNA Quantification & Quality Assessment - Geoduck & Oly gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
DNA Isolations – Oly Fidalgo 2SN Ctenidia
2015
Olympia oyster reciprocal transplant
DNA Isolation – Geoduck & Olympia Oyster
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Adaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Restriction Digest – Oly gDNA for RAD-seq w/AlfI
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Troubleshooting - Oly RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
PCR - Oly RAD-seq Prep Scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
PCR - Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Adaptor Ligation – Oly AlfI-Digested gDNA for RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Restriction Digest – Oly gDNA for RAD-seq w/AlfI
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Sample Submission - Additional Olympia Oyster gDNA for Genome Sequencing @ BGI
2015
Olympia Oyster Genome Sequencing
Samples Submitted
Agarose Gel - Geoduck & Olympia Oyster gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Olympia oyster reciprocal transplant
DNA Quantification - Pooled geoduck gDNA
2015
Geoduck Genome Sequencing
DNA Isolation - Geoduck & Olympia Oyster
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Olympia oyster reciprocal transplant
PCR - Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
PCR - Oly RAD-seq Test-scale PCR
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Reagent Prep
Adaptor Ligation - Oly AlfI-Digested gDNA for RAD-seq
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Reagent Prep
Restriction Digest - Oly gDNA for RAD-seq w/AlfI
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Reagent Prep
Sample Submission - Olympia Oyster gDNA for Genome Sequencing @ BGI
2015
Olympia Oyster Genome Sequencing
Samples Submitted
Agarose Gel - Geoduck & Olympia oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Ethanol Precipitation - Geoduck & Olympia oyster gDNA
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Agarose Gel - Olympia oyster Whole Body gDNA Integrity Check
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
DNA Isolation - Olympia oyster whole body
2015
2bRAD Library Tests for Sequencing at Genewiz
Olympia oyster reciprocal transplant
Agarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Agarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
Agarose Gel - Geoduck & Olympia Oyster gDNA Integrity Check
2015
Geoduck Genome Sequencing
Olympia Oyster Genome Sequencing
RAD-Seq Library Prep Reagents
2015
2bRAD Library Tests for Sequencing at Genewiz
Reagent Prep
RNA Quantification - O.lurida 1hr post-mechanical heat stress DNased RNA
2015
Olympia oyster reciprocal transplant
RNA Isolation – O.lurida Ctenidia 1hr Post-Mechanical Stress
2015
Olympia oyster reciprocal transplant
RNA Isolation - O.lurida Ctenidia 1hr Post-Mechanical Stress
2015
Olympia oyster reciprocal transplant
RNAseq Data Receipt - Geoduck Gonad RNA 100bp PE Illumina
2015
Protein expression profiles during sexual maturation in Geoduck
Sample Submission - Geoduck Gonad for RNA-seq
2015
Protein expression profiles during sexual maturation in Geoduck
Bioanalyzer - Geoduck Gonad RNA Quality Assessment
2015
Protein expression profiles during sexual maturation in Geoduck
Bioinformatics – Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Bioinformatics - Trimmomatic/FASTQC on C.gigas Larvae OA NGS Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
BLAST - C.gigas Larvae OA Illumina Data Against GenBank nt DB
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Goals - May 2015
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Goals
Lineage-specific DNA methylation patterns in developing oysters
LSU C.virginica Oil Spill MBD BS Sequencing
Protein expression profiles during sexual maturation in Geoduck
BLASTN - C.gigas OA Larvae to C.gigas Ensembl 1.24 BLAST DB
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Bioanalyzer Data - Geoduck RNA from Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
RNA Isolation – Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Bioanalyzer Submission - Geoduck Gonad RNA from Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Quality Trimming - C.gigas Larvae OA BS-Seq Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Quality Trimming - LSU C.virginica Oil Spill MBD BS-Seq Data
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Sequence Data Analysis - LSU C.virginica Oil Spill MBD BS-Seq Data
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Sequence Data Analysis - C.gigas Larvae OA BS-Seq Data
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Sequence Data - C.gigas OA Larvae BS-Seq Demultiplexed
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
RNA Isolation - Geoduck Gonad in Paraffin Histology Blocks
2015
Protein expression profiles during sexual maturation in Geoduck
Sequencing Data - C.gigas Larvae OA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
TruSeq Adaptor Counts – LSU C.virginica Oil Spill Sequences
2015
LSU C.virginica Oil Spill MBD BS Sequencing
TruSeq Adaptor Identification Method Comparison - LSU C.virginica Oil Spill Sequences
2015
LSU C.virginica Oil Spill MBD BS Sequencing
DNA Quantification - Claire’s C.gigas Sheared DNA
2015
Lineage-specific DNA methylation patterns in developing oysters
Library Quality Assessment - C.gigas OA larvae Illumina libraries
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
BS-seq Library Prep - C.gigas Larvae OA 1000ppm
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
DNA Quantification - C.gigas Larvae 1000ppm
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
DNA Quantification - Claire’s Sheared C.gigas Mantle Heat Shock Samples
2015
Lineage-specific DNA methylation patterns in developing oysters
Bioanalyzer - C.gigas Sheared DNA from 20140108
2015
Lineage-specific DNA methylation patterns in developing oysters
Library Prep - Quantification of C.gigas larvae OA 1000ppm library
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Sequencing Data - LSU C.virginica MBD BS-Seq
2015
LSU C.virginica Oil Spill MBD BS Sequencing
Bisulfite NGS Library Prep - Bisulfite Conversion & Illumina Library Construction of C.gigas larvae DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Bisuflite NGS Library Prep – C.gigas larvae OA bisulfite library quantification
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Bisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA (continued from yesterday)
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Bisuflite NGS Library Prep - C.gigas larvae OA bisulfite DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
DNA Bisulfite Conversion - C.gigas larvae OA Sheared DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
SpeedVac - C.gigas larvae OA DNA
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
DNA Isolation - C.gigas larvae from 2011 NOAA OA Experiment
2015
Crassostrea gigas larvae OA (2011) bisulfite sequencing
Bisulfite NGS Library Prep - LSU C.virginica Oil Spill MBD Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA (continued from yesterday)
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
LSU C.virginica Oil Spill MBD BS Sequencing
Bisulfite NGS Library Prep - LSU C.virginica Oil Spill Bisulfite DNA and Emma’s C.gigas Larvae OA Bisulfite DNA
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
LSU C.virginica Oil Spill MBD BS Sequencing
Bisulfite Conversion - LSU C.virginica Oil Spill MBD DNA and Emma’s C.gigas Larvae OA DNA
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
LSU C.virginica Oil Spill MBD BS Sequencing
DNA Isolation - Claire’s C.gigas Female Gonad for Illumina Bisulfite Sequencing
2014
Lineage-specific DNA methylation patterns in developing oysters
Gel - Sheared gDNA
2014
LSU C.virginica Oil Spill MBD BS Sequencing
DNA Isolation - C.gigas Larvae from Emma OA Experiments
2014
Crassostrea gigas larvae OA (2011) bisulfite sequencing
RAD Sequencing - Oly Oyster gDNA-01 RAD Library (from 20141110)
2014
Olympia oyster reciprocal transplant
Samples Submitted
Library Prep - Oly Oyster gDNA-01 RAD
2014
Olympia oyster reciprocal transplant
Ligation - Illumina P1 Adapters for Oly Oyster gDNA-01 RAD Sequencing (from 20141031)
2014
Olympia oyster reciprocal transplant
Restriction Digest - Oly Oyster gDNA-01 for RAD Sequencing (from 20141029)
2014
Olympia oyster reciprocal transplant
DNA Isolation - Claire’s C.gigas Female Gonad
2014
Lineage-specific DNA methylation patterns in developing oysters
DNA Isolation - Test Sample
2014
Lineage-specific DNA methylation patterns in developing oysters
Miscellaneous
Phenol-Chloroform DNA Clean Up - Mac and Claire’s Samples (from 20140410)
2014
Lineage-specific DNA methylation patterns in developing oysters
DNA Isolation - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad
2014
Lineage-specific DNA methylation patterns in developing oysters
DNA gel - Claire’s C.gigas Female Gonad and Mac’s C.gigas Gonad
2014
Lineage-specific DNA methylation patterns in developing oysters
DNA Isolation - C.gigas Female Gonads (from frozen)
2014
Lineage-specific DNA methylation patterns in developing oysters
Sample Submission - QPX RNA and DNA for Illumina 36bp single-end RNA/DNAseq
2012
Miscellaneous
Samples Submitted
Reverse Transcription - DNased C.gigas Larval RNA from 20120427
2012
Miscellaneous
M-MLV
reverse transcription
RNA
Vibrio tubiashii
qPCR - Hard Clam Primers on cDNA from yesterday
2010
Miscellaneous
BigDef
C1qTNG
CA
CFX96
CytP450-like
ferritnin
graphs
GST
Hard clam
HemoDef
HSP 70
Immomix
lysozyme
MA
MAX
Mercenaria mercenaria
Mercenaria_Rel
metallothionein
Mm_TRAF6
NADHIV
qPCR
RACK1
SenescentProt
STI
SYTO 13
TfAP1
TLR
gDNA Isolation - Mac gigas gill samples
2010
Miscellaneous
Crassostrea gigas
DNA Isolation
DNazol
gill
Pacific oyster
R37
R51
RNAlater
Package Rec’d - From NOAA in Connecticut
2010
Miscellaneous
algae
Isochrysis sp. T-150
NOAA
package
Tetraselmis cheri Ply429
Thalassiaosira weissflugii TW
gDNA Isolation - Mac gigas gill samples
2010
Miscellaneous
Crassostrea gigas
DNazol
Pacific oyster
R51
RNAlater
DNA Isolation - Qiagen Kit Comparison
2010
Miscellaneous
DNeasy
gDNA
gel
Hyperladder I
troubleshooting
PCR - Bay/Sea scallop gDNA isolated earlier today
2009
Miscellaneous
SDS/PAGE, Western Blot - Test of new Western Breeze Kit & HSP70 Ab for FISH441
2008
Miscellaneous
anti-HSP70
anti-myc
antibody
Coomassie
Crassostrea gigas
MSTN
MSTN1b
myostatin
Pacific oyster
protein
SDS-PAGE
SeeBlue Plus
Western blot
Western Breeze Chromogenic (anti-mouse) Kit
RNA Isolation - V. tubiashii from challenge (see 20081216)
2008
Miscellaneous
MICROBExpress Kit
NanoDrop1000
RNA isolation
RNA quantification
TriReagent
Vibrio tubiashii
Vibrio challenge
2008
Miscellaneous
bacterial challenge
bacterial culture
Vibrio exposure
Vibrio tubiashii
SDS/PAGE/Western - anti-HSP70 Ab Re-test
2008
Miscellaneous
anti-HSP70
antibody
chemiluminescent
Coomassie
Crassostrea gigas
hemocyte
hemolymph
Pacific oyster
protein
SDS-PAGE
SeeBlue Plus
SDS/PAGE/Western - anti-HSP70 Ab test
2008
Miscellaneous
anti-HSP70
antibody
Coomassie
Crassostrea gigas
gill
mucus
Octopus rubescans
Pacific oyster
protein
red octopus
SDS-PAGE
skin
Sequencing - Y2H colony PCRs from 20081112
2008
Myostatin Interacting Proteins
No matching items
-sRNA-seq-Data-from-Azenta-Project-30-1035633055/20241001-rphi-rSNAseq-checksums-01.png)